-
 RNA Sequencing and Weighted Gene Co-Expression Network Analysis Highlight DNA Replication and Key Genes in Nucleolin-Depleted Hepatoblastoma Cells
RNA Sequencing and Weighted Gene Co-Expression Network Analysis Highlight DNA Replication and Key Genes in Nucleolin-Depleted Hepatoblastoma Cells -
 An Updated Analysis of Exon-Skipping Applicability for Duchenne Muscular Dystrophy Using the UMD-DMD Database
An Updated Analysis of Exon-Skipping Applicability for Duchenne Muscular Dystrophy Using the UMD-DMD Database -
 Update on Inherited Pediatric Motor Neuron Diseases: Clinical Features and Outcome
Update on Inherited Pediatric Motor Neuron Diseases: Clinical Features and Outcome
Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Biochemistry and Molecular Biology (SEBBM) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics and Heredity) / CiteScore - Q2 (Genetics (clinical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 14.9 days after submission; acceptance to publication is undertaken in 2.6 days (median values for papers published in this journal in the second half of 2024).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
2.8 (2023);
5-Year Impact Factor:
3.3 (2023)
Latest Articles
A Genome-Wide Association Study of First-Episode Psychosis: A Genetic Exploration in an Italian Cohort
Genes 2025, 16(4), 439; https://doi.org/10.3390/genes16040439 (registering DOI) - 7 Apr 2025
Abstract
Background: Psychosis, particularly schizophrenia (SZ), is influenced by genetic and environmental factors. The neurodevelopmental hypothesis suggests that genetic factors affect neuronal circuit connectivity during perinatal periods, hence causing the onset of the diseases. In this study, we performed a genome-wide association study (GWAS)
[...] Read more.
Background: Psychosis, particularly schizophrenia (SZ), is influenced by genetic and environmental factors. The neurodevelopmental hypothesis suggests that genetic factors affect neuronal circuit connectivity during perinatal periods, hence causing the onset of the diseases. In this study, we performed a genome-wide association study (GWAS) in a sample of the first episode of psychosis (FEP). Methods: A sample of 147 individuals diagnosed with non-affective psychosis and 102 controls were recruited and assessed. After venous blood and DNA extraction, the samples were genotyped. Genetic data underwent quality controls, genotype imputation, and a case-control genome-wide association study (GWAS). After the GWAS, results were investigated using an in silico functional mapping and annotation approach. Results: Our GWAS showed the association of 27 variants across 13 chromosomes at genome-wide significance (p < 1 × 10−7) and a total of 1976 candidate variants across 188 genes at suggestive significance (p < 1 × 10−5), mostly mapping in non-coding or intergenic regions. Gene-based tests reported the association of the SUFU (p = 4.8 × 10−7) and NCAN (p = 1.6 × 10−5) genes. Gene-sets enrichment analyses showed associations in the early stages of life, spanning from 12 to 24 post-conception weeks (p < 1.4 × 10−3) and in the late prenatal period (p = 1.4 × 10−3), in favor of the neurodevelopmental hypothesis. Moreover, several matches with the GWAS Catalog reported associations with strictly related traits, such as SZ, as well as with autism spectrum disorder, which shares some genetic overlap, and risk factors, such as neuroticism and alcohol dependence. Conclusions: The resulting genetic associations and the consequent functional analysis displayed common genetic liability between the non-affective psychosis, related traits, and risk factors. In sum, our investigation provided novel hints supporting the neurodevelopmental hypothesis in SZ and—in general—in non-affective psychoses.
Full article
(This article belongs to the Special Issue Genetics and Genomics of Psychiatric Disorders)
►
Show Figures
Open AccessArticle
Overexpression of FLZ12 Suppresses Root Hair Development and Enhances Iron-Deficiency Tolerance in Arabidopsis
by
Mingke Yan, Xin Zhang and Jinghui Gao
Genes 2025, 16(4), 438; https://doi.org/10.3390/genes16040438 (registering DOI) - 6 Apr 2025
Abstract
Background: The Arabidopsis FCS-LIKE ZINC FINGER (FLZ) family proteins play crucial roles in responses to various biotic and abiotic stresses, but the functions of many family members remain uncharacterized. Methods: In this study, we investigated the function of FLZ12, a member
[...] Read more.
Background: The Arabidopsis FCS-LIKE ZINC FINGER (FLZ) family proteins play crucial roles in responses to various biotic and abiotic stresses, but the functions of many family members remain uncharacterized. Methods: In this study, we investigated the function of FLZ12, a member of the FLZ family, using a reverse genetic approach. Results: We found that overexpression of FLZ12 impaired root hair development, as evidenced by marked reductions in both root hair length and number under normal growth conditions. However, deprivation of phosphate could partially restore root hair formation, although it still impeded root hair elongation. Notably, FLZ12-overexpressing lines exhibited greatly enhanced tolerance to iron deficiency, with seedlings exhibiting more vigorous and robust growth compared to wild-type plants. In contrast, knockout of FLZ12 resulted in slight impact on seedling development. Further analysis revealed that FLZ12 accumulation was increased in vascular tissues of plants subjected to iron starvation, and the protein was predominantly localized within the nucleus. Conclusions: Integrating these findings with existing evidence, we propose that FLZ12 functions as a translational regulator through interacting with other proteins, playing dual roles in root hair development and iron-deficiency responses in Arabidopsis. These findings provide new insights into the FLZ-domain-containing proteins and offer molecular strategies to enhance iron uptake efficiency in crops, highlighting FLZ12 as a promising candidate for future breeding efforts.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
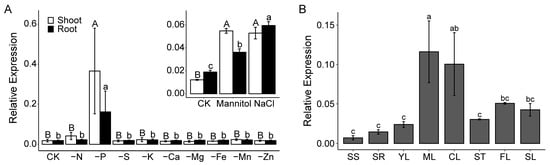
Figure 1
Open AccessReview
RON Receptor Signaling and the Tumor Microenvironment
by
Emily Wachter, Levi H. Fox, Zhixin Lu, Angelle D. Jones, Nicholas D. Casto and Susan E. Waltz
Genes 2025, 16(4), 437; https://doi.org/10.3390/genes16040437 (registering DOI) - 6 Apr 2025
Abstract
The immune microenvironment plays a critical role in tumor growth and development. Immune activation within the tumor microenvironment is dynamic and can be modulated by tumor intrinsic and extrinsic signaling. The RON receptor tyrosine kinase is canonically associated with growth signaling and wound
[...] Read more.
The immune microenvironment plays a critical role in tumor growth and development. Immune activation within the tumor microenvironment is dynamic and can be modulated by tumor intrinsic and extrinsic signaling. The RON receptor tyrosine kinase is canonically associated with growth signaling and wound healing, and this receptor is frequently overexpressed in a variety of cancers. Epithelial cells, macrophages, dendritic cells, and fibroblasts express RON, presenting an important axis by which RON overexpressing tumors influence the tumor microenvironment. This review synthesizes the existing literature on the roles of tumor cell-intrinsic and -extrinsic RON signaling, highlighting areas of interest and gaps in knowledge that show potential for future studies.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
A Knockout of the OsGAPDHC6 Gene Encoding a Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase Reacts Sensitively to Abiotic Stress in Rice
by
Jin-Young Kim, Ye-Ji Lee, Hye-Mi Lee, Yoo-Seob Jung, Jiyun Go, Hyo-Ju Lee, Ki-Sun Nam, Jong-Hee Kim, Kwon-Kyoo Kang and Yu-Jin Jung
Genes 2025, 16(4), 436; https://doi.org/10.3390/genes16040436 (registering DOI) - 6 Apr 2025
Abstract
Background/Objectives: The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzyme, encoded by OsGAPDHC6, plays a crucial role in glycolysis while participating in various physiological and stress response pathways. Methods: In this study, the expression levels of the OsGAPDHC1 and OsGAPDHC6 genes were investigated over time by
[...] Read more.
Background/Objectives: The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) enzyme, encoded by OsGAPDHC6, plays a crucial role in glycolysis while participating in various physiological and stress response pathways. Methods: In this study, the expression levels of the OsGAPDHC1 and OsGAPDHC6 genes were investigated over time by treating various abiotic stresses (ABA, PEG, NaCl, heat, and cold) in rice seedlings. Results: As a result, the expression levels of both genes in the ABA-treated group increased continuously for 0–6 h and then de-creased sharply from 12 h onwards. The mutational induction of the GAPDHC6 gene by the CRISPR/Cas9 system generated a stop codon through a 1 bp insertion into protein production. The knockout (KO) lines showed differences in seed length, seed width, and seed thickness compared to wild-type (WT) varieties. In addition, KO lines showed a lower germination rate, germination ability, and germination index of seeds under salt treatment compared to WT, and leaf damage due to 3,3′-diaminobenzidine (DAB) staining was very high due to malondialdehyde (MDA) accumulation. The KO line was lower regarding the expression level of stress-related genes compared to WT. Conclusions: Therefore, the OsGAPDHC6 gene is evaluated as a gene that can increase salt resistance in rice as it actively responds to salt stress in the early stages of growth, occurring from seed germination to just before the tilling stage.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Transcriptomic Characterization of miRNAs in Pyrrhalta aenescens Fairmaire in Response to 20-Hydroxyecdysone Treatment
by
Jie Liu, Li Gao, Chao Du, Tianfeng Duan and Li Liu
Genes 2025, 16(4), 435; https://doi.org/10.3390/genes16040435 (registering DOI) - 5 Apr 2025
Abstract
Background/Objectives: Pyrrhalta aenescens, a major pest of elm trees, causes extensive ecological and economic damage through rapid population growth and defoliation. Existing research mainly focuses on its biological traits and chemical control, with little knowledge about its reproductive development mechanisms, a key
[...] Read more.
Background/Objectives: Pyrrhalta aenescens, a major pest of elm trees, causes extensive ecological and economic damage through rapid population growth and defoliation. Existing research mainly focuses on its biological traits and chemical control, with little knowledge about its reproductive development mechanisms, a key factor in population expansion. In other insects, the steroid hormone 20-hydroxyecdysone (20E) regulates development and reproduction via microRNA (miRNA)-mediated pathways, but this has not been studied in P. aenescens. This study aimed to systematically identify miRNAs responsive to 20E in P. aenescens and unravel their roles in regulating reproduction and metabolic pathways, providing foundational insights into hormone–miRNA crosstalk in this ecologically significant pest. Methods: Adult beetles (collected from Baotou, Inner Mongolia) were injected with 1.0 μg/μL 20E or control. Total RNA from three biological replicates (10 adults each) was sequenced, followed by miRNA identification, differential expression analysis, target prediction, and functional enrichment. Results: Small RNA sequencing identified 205 miRNAs (162 conserved, 43 novel), with 12 DEMs post-20E treatment. Target prediction linked these miRNAs to 7270 genes, including key regulators of the FoxO signaling pathway and MAPK signaling pathway. KEGG analysis highlighted lipid metabolism and stress response pathways. Conclusions: This study revealed that 20E modulates miRNA networks to regulate FoxO and MAPK pathways in P. aenescens, suggesting hormonal control of lipid metabolism and developmental processes. As the first miRNA resource for this pest, our findings provide mechanistic insights into 20E–miRNA crosstalk and identify potential molecular targets for disrupting its reproductive biology, laying a foundation for eco-friendly pest control.
Full article
(This article belongs to the Special Issue Genomics, Transcriptomics, and Proteomics of Insects)
►▼
Show Figures

Figure 1
Open AccessCase Report
Maternal Uniparental Isodisomy of Chromosome 6: A Novel Case of Teratoma and Autism Spectrum Disorder with a Diagnostic and Management Framework
by
Aleksandra Świeca, Maria Franaszczyk, Agnieszka Maryniak, Patryk Lipiński, Rafał Płoski and Krzysztof Szczałuba
Genes 2025, 16(4), 434; https://doi.org/10.3390/genes16040434 (registering DOI) - 5 Apr 2025
Abstract
Background: Uniparental disomy (UPD) is the inheritance of both copies of a chromosome from a single parent, leading to distinct genetic conditions. Maternal UPD of chromosome 6 (UPD(6)mat) is extremely rare, with few molecularly confirmed cases reported. Methods: We report a prematurely born
[...] Read more.
Background: Uniparental disomy (UPD) is the inheritance of both copies of a chromosome from a single parent, leading to distinct genetic conditions. Maternal UPD of chromosome 6 (UPD(6)mat) is extremely rare, with few molecularly confirmed cases reported. Methods: We report a prematurely born female with isodisomic UPD(6)mat, presenting with intrauterine growth restriction (IUGR), developmental delay, autism spectrum disorder, dysmorphic features, and a sacrococcygeal teratoma. In addition, we reviewed 24 confirmed UPD(6)mat cases to assess clinical patterns in prenatal findings, birth outcomes, and postnatal features. Results: Trio whole-exome sequencing revealed complete isodisomy of chromosome 6 and a de novo heterozygous DIAPH2 variant of uncertain significance. In the literature review, IUGR was present in 87% of cases, with most individuals born small for gestational age and preterm. Failure to thrive and neurodevelopmental issues were also frequent. While the exact molecular basis remains unknown, imprinting disturbances—similar to those in UPD(6)pat—and cryptic trisomy 6 mosaicism, particularly in heterodisomy, are the most likely mechanisms. No specific gene or consistent epigenetic abnormality has been identified. Conclusions: This study aims to enhance the understanding of the genetic and phenotypic spectrum of UPD(6)mat, improving diagnostic and management approaches for this ultra-rare genetic disorder. We propose a detailed list of clinical assessments and tests to be performed following the detection of maternal uniparental disomy of chromosome 6.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures
Figure 1
Open AccessArticle
Exploring the Dietary Strategies of Coated Sodium Butyrate: Improving Antioxidant Capacity, Meat Quality, Fatty Acid Composition, and Gut Health in Broilers
by
Zhuoya Gu, Wenwu Xu, Tiantian Gu, Lizhi Lu and Guohong Chen
Genes 2025, 16(4), 433; https://doi.org/10.3390/genes16040433 (registering DOI) - 5 Apr 2025
Abstract
Background/Objectives: Broiler chickens are excellent animals for protein production and play an essential role in the food industry. The purpose of this study is to investigate the effect of coated sodium butyrate (CSB) on the biochemical indices, antioxidant capacity, meat quality, fatty acid
[...] Read more.
Background/Objectives: Broiler chickens are excellent animals for protein production and play an essential role in the food industry. The purpose of this study is to investigate the effect of coated sodium butyrate (CSB) on the biochemical indices, antioxidant capacity, meat quality, fatty acid composition, and gut health of Xianju broilers. Methods: A total of 192 one-day-old broilers were randomly divided into four treatment groups: the basal diet (CK), the basal diet with 250 mg/kg CSB (CSB250), the basal diet with 500 mg/kg CSB500 (CSB500), and the basal diet with 1000 mg/kg CSB (CSB1000). Each group included six replicates, with eight chicks per replicate. Results: We found that CSB supplementation in the diets has no function on plasma biochemical indices; however, CSB1000 broilers exhibited markedly elevated plasma TG levels. Furthermore, CSB supplementation at different concentrations significantly increased plasma antioxidase capacity in broilers. Moreover, breast meat supplemented with CSB displayed a higher shear force, pH24h, and inosinic acid content than CK meat. Breast meat of broilers fed CSB1000 showed improved fatty acid composition, evidenced by increased levels of polyunsaturated fatty acids (C16:1, C18:2, C22:4, and C22:6). Moreover, supplementation with CSB1000 optimized the gut microbiota composition, particularly by enhancing the abundance of Firmicutes and the Firmicutes/Bacteroidetes ratio. Conclusions: Collectively, these findings offer a basis for the extensive application of CSB as a feed addition to enhance the quality of meat in the broiler sector.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
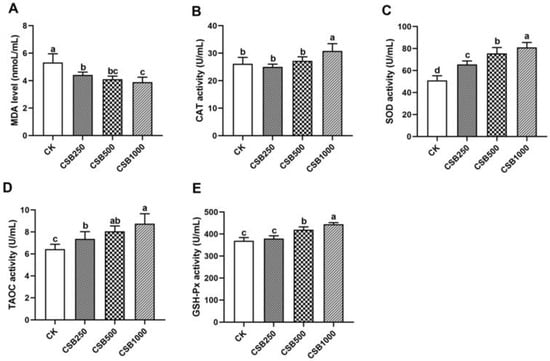
Figure 1
Open AccessArticle
GC Content in Nuclear-Encoded Genes and Effective Number of Codons (ENC) Are Positively Correlated in AT-Rich Species and Negatively Correlated in GC-Rich Species
by
Douglas M. Ruden
Genes 2025, 16(4), 432; https://doi.org/10.3390/genes16040432 (registering DOI) - 5 Apr 2025
Abstract
Background/Objectives: Codon usage bias affects gene expression and translation efficiency across species. The effective number of codons (ENC) and GC content influence codon preference, often displaying unimodal or bimodal distributions. This study investigates the correlation between ENC and GC rankings across species and
[...] Read more.
Background/Objectives: Codon usage bias affects gene expression and translation efficiency across species. The effective number of codons (ENC) and GC content influence codon preference, often displaying unimodal or bimodal distributions. This study investigates the correlation between ENC and GC rankings across species and how their relationship affects codon usage distributions. Methods: I analyzed nuclear-encoded genes from 17 species representing six kingdoms: one bacteria (Escherichia coli), three fungi (Saccharomyces cerevisiae, Neurospora crassa, and Schizosaccharomyces pombe), one archaea (Methanococcus aeolicus), three protists (Rickettsia hoogstraalii, Dictyostelium discoideum, and Plasmodium falciparum),), three plants (Musa acuminata, Oryza sativa, and Arabidopsis thaliana), and six animals (Anopheles gambiae, Apis mellifera, Polistes canadensis, Mus musculus, Homo sapiens, and Takifugu rubripes). Genes in all 17 species were ranked by GC content and ENC, and correlations were assessed. I examined how adding or subtracting these rankings influenced their overall distribution in a new method that I call Two-Rank Order Normalization or TRON. The equation, TRON = SUM(ABS((GC rank1:GC rankN) − (ENC rank1:ENC rankN))/(N2/3), where (GC rank1:GC rankN) is a rank-order series of GC rank, (ENC rank1:ENC rankN) is a rank-order series ENC rank, sorted by the rank-order series GC rank. The denominator of TRON, N2/3, is the normalization factor because it is the expected value of the sum of the absolute value of GC rank–ENC rank for all genes if GC rank and ENC rank are not correlated. Results: ENC and GC rankings are positively correlated (i.e., ENC increases as GC increases) in AT-rich species such as honeybees (R2 = 0.60, slope = 0.78) and wasps (R2 = 0.52, slope = 0.72) and negatively correlated (i.e., ENC decreases as GC increases) in GC-rich species such as humans (R2 = 0.38, slope = −0.61) and rice (R2 = 0.59, slope = −0.77). Second, the GC rank–ENC rank distributions change from unimodal to bimodal as GC content increases in the 17 species. Third, the GC rank+ENC rank distributions change from bimodal to unimodal as GC content increases in the 17 species. Fourth, the slopes of the correlations (GC versus ENC) in all 17 species are negatively correlated with TRON (R2 = 0.98) (see Graphic Abstract). Conclusions: The correlation between ENC rank and GC rank differs among species, shaping codon usage distributions in opposite ways depending on whether a species’ nuclear-encoded genes are AT-rich or GC-rich. Understanding these patterns might provide insights into translation efficiency, epigenetics mediated by CpG DNA methylation, epitranscriptomics of RNA modifications, RNA secondary structures, evolutionary pressures, and potential applications in genetic engineering and biotechnology.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Molecular Signatures of Exercise Adaptation in Arabian Racing Horses: Transcriptomic Insights from Blood and Muscle
by
Monika Stefaniuk-Szmukier, Tomasz Szmatoła and Katarzyna Ropka-Molik
Genes 2025, 16(4), 431; https://doi.org/10.3390/genes16040431 - 4 Apr 2025
Abstract
Background/Objectives: Human-driven selection has shaped modern horse breeds into highly specialized athletes, particularly in racing. Arabian horses, renowned for their endurance, provide an excellent model for studying molecular adaptations to exercise. This study aimed to identify genes commonly influenced by physical exertion in
[...] Read more.
Background/Objectives: Human-driven selection has shaped modern horse breeds into highly specialized athletes, particularly in racing. Arabian horses, renowned for their endurance, provide an excellent model for studying molecular adaptations to exercise. This study aimed to identify genes commonly influenced by physical exertion in the gluteus medius muscle and whole blood of Arabian horses during their first year of race training. Methods: RNA sequencing of sixteen pure-breed Arabian horses was used to analyze transcriptomic changes at three key training stages. Differentially expressed genes (DEGs) were identified to explore their role in endurance and metabolic adaptation. Results: Seven genes—RCHY1, PIH1D1, IVD, FABP3, ANKRD2, USP13, and CRYAB—were consistently deregulated across tissues and training periods. These genes are involved in muscle remodeling, metabolism, oxidative stress response, and protein turnover. ANKRD2 was associated with mechanosensing and muscle adaptation, FABP3 with fatty acid metabolism, and USP13 with ubiquitination-related pathways crucial for muscle recovery and energy regulation. The transcriptomic overlap between muscle and blood suggests potential systemic biomarkers for athletic performance and endurance. Conclusions: Our findings highlight the importance of multi-tissue transcriptomic profiling in understanding exercise-induced molecular adaptations. The identified genes warrant further investigation as potential molecular markers for monitoring training progression and athletic potential in endurance horses. This study contributes to the growing field of equine sports genetics and may offer translational insights into human sports performance.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
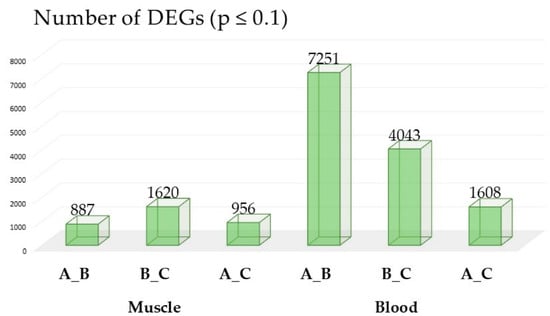
Figure 1
Open AccessArticle
Abundant β-Defensin Copy Number Variations in Pigs
by
Dohun Kim, Hye-sun Cho, Mingue Kang, Byeongyong Ahn, Jaeyeol Shin and Chankyu Park
Genes 2025, 16(4), 430; https://doi.org/10.3390/genes16040430 - 4 Apr 2025
Abstract
Background/Objectives: β-defensins are a family of classical endogenous antimicrobial peptides involved in innate immune response. β-defensins are encoded by a large number of loci and known to show extensive copy number variations (CNVs) that may be useful as DNA markers for host resilience
[...] Read more.
Background/Objectives: β-defensins are a family of classical endogenous antimicrobial peptides involved in innate immune response. β-defensins are encoded by a large number of loci and known to show extensive copy number variations (CNVs) that may be useful as DNA markers for host resilience against pathogenic infections. Methods: We developed a quantitative PCR-based method to estimate the genomic copy numbers of 13 pig β-defensin (pBD) genes and analyzed the range and extent of CNVs across several commercial pig breeds. Results: We assessed 38 animals from four pure breeds and a crossbreed and observed CNVs ranging from two to five genomic copies from pBD114, pBD115, pBD119, pBD124, pBD128, and pBD129, indicating extensive individual variations of gene copy numbers of these genes within each breed. The mean copy numbers of these pBDs were lower in Landrace and higher in Berkshire than in other breeds. We also observed a strong correlation between the genomic copy number and their expression levels with the correlation coefficient (r) > 0.9 for pBD114, pBD119, and pBD129 in the kidney, with these genes being highly expressed. Conclusions: Although we only analyzed 13 pBDs among 29 reported genes, our results showed the presence of extensive CNVs in β-defensins from pigs. The genomic copy number of β-defensins may contribute to improving animal resilience against pathogenic infections and other associated phenotypes.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
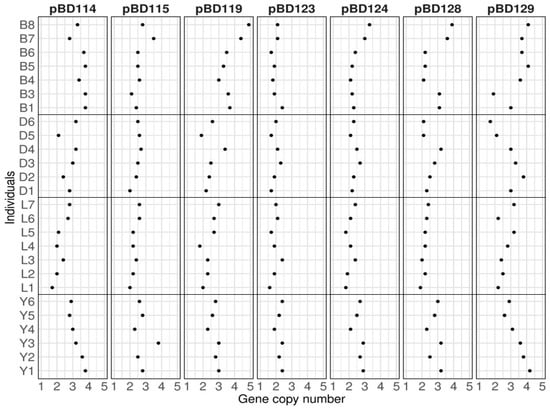
Figure 1
Open AccessArticle
Identification and Expression Analysis of CCCH Zinc Finger Family Genes in Oryza sativa
by
Zhihan Wang, Shunyuan Li, Hongkai Wu, Linzhou Huang, Liangbo Fu, Chengfang Zhan, Xueli Lu, Long Yang, Liping Dai and Dali Zeng
Genes 2025, 16(4), 429; https://doi.org/10.3390/genes16040429 - 3 Apr 2025
Abstract
Background: CCCH zinc finger proteins (OsC3Hs) are a class of transcriptional regulators that play important roles in plant development and stress responses. Although their functional significance has been widely studied in model species, comprehensive genome-wide characterization of CCCH proteins in rice (Oryza
[...] Read more.
Background: CCCH zinc finger proteins (OsC3Hs) are a class of transcriptional regulators that play important roles in plant development and stress responses. Although their functional significance has been widely studied in model species, comprehensive genome-wide characterization of CCCH proteins in rice (Oryza sativa) remains limited. Methods: Using Arabidopsis CCCH proteins as references, we identified the CCCH gene family in rice and analyzed the physicochemical properties, subcellular localization, conserved structures, phylogeny, cis-regulatory elements, synteny analysis, spatiotemporal expression patterns, and expression patterns under drought, ABA, and MeJA treatments for the identified CCCH family members. Results: The results showed that the rice CCCH family comprises 73 members, which are unevenly distributed across the 12 chromosomes. Phylogenetic analysis classified them into 11 subfamilies. Subcellular localization indicated that most members are localized in the nucleus. The upstream regions of CCCH promoters contain a large number of cis-regulatory elements related to plant hormones and biotic stress responses. Most genes respond to drought, abscisic acid (ABA), and methyl jasmonate (MeJA) treatments. OsC3H36 was highly expressed under drought, ABA, and MeJA treatments. Haplotype analysis of this gene revealed two major allelic variants (H1 and H2), with H1 predominantly found in japonica rice and associated with increased grain width and 1000-grain weight. Functional validation using a chromosome segment substitution line (CSSL1) confirmed these findings. Conclusions: CCCH genes play important roles in rice growth, development, and stress responses. Additionally, we validated that OsC3H36 is associated with rice grain width and 1000-grain weight.
Full article
(This article belongs to the Special Issue Genetics and Breeding of Rice)
►▼
Show Figures

Figure 1
Open AccessFeature PaperReview
Genetics of Suicide
by
Mostafa Khalil, Anil Kalyoncu and Alfredo Bellon
Genes 2025, 16(4), 428; https://doi.org/10.3390/genes16040428 - 3 Apr 2025
Abstract
Over the past two decades, suicide has consistently ranked among the leading causes of death in the United States. While suicide deaths are closely associated with uicidal ideation and attempts, these are not good predictors of future suicide deaths. Establishing who is at
[...] Read more.
Over the past two decades, suicide has consistently ranked among the leading causes of death in the United States. While suicide deaths are closely associated with uicidal ideation and attempts, these are not good predictors of future suicide deaths. Establishing who is at risk of suicide remains a challenge that is mostly hampered by the lack of understanding of its pathophysiology. Nonetheless, evidence continues to accumulate suggesting that suicide is driven by a complex and dynamic interaction between environmental factors and genetics. The identification of genes that place people at risk of suicide remains elusive, but data are rapidly evolving. In this narrative review, we describe how Tryptophan hydroxylase (TPH) genes, particularly TPH1 and TPH2, have been associated with suicide in various publications. There is also replicated evidence linking the brain-derived neurotrophic factor gene to suicide, with its most consistent results originating from epigenetic studies. Not surprisingly, many genes involved in the hypothalamic–pituitary–adrenal axis have been connected with suicide, but these data require replication. Finally, among the inflammatory genes studied in suicide, only specific polymorphisms in TNF-alpha and IL-6 may increase susceptibility to suicidal behavior. In conclusion, significant work remains to be performed as inconsistencies undermine the reliability of genetic results in suicide. Potential avenues for future research are proposed.
Full article
Open AccessArticle
Advancing Non-Invasive Prenatal Screening: A Targeted 1069-Gene Panel for Comprehensive Detection of Monogenic Disorders and Copy Number Variations
by
Roberto Sirica, Alessandro Ottaiano, Luigi D’Amore, Monica Ianniello, Nadia Petrillo, Raffaella Ruggiero, Rosa Castiello, Alessio Mori, Eloisa Evangelista, Luigia De Falco, Mariachiara Santorsola, Michele Misasi, Giovanni Savarese and Antonio Fico
Genes 2025, 16(4), 427; https://doi.org/10.3390/genes16040427 - 2 Apr 2025
Abstract
We introduce an innovative, non-invasive prenatal screening approach for detecting fetal monogenic alterations and copy number variations (CNVs) from maternal blood. Method: Circulating free DNA (cfDNA) was extracted from maternal peripheral blood and processed using the VeriSeq NIPT Solution (Illumina, San Diego, CA,
[...] Read more.
We introduce an innovative, non-invasive prenatal screening approach for detecting fetal monogenic alterations and copy number variations (CNVs) from maternal blood. Method: Circulating free DNA (cfDNA) was extracted from maternal peripheral blood and processed using the VeriSeq NIPT Solution (Illumina, San Diego, CA, USA), with shallow whole-genome sequencing (sWGS) performed on a NextSeq550Dx (Illumina). A customized gene panel and bioinformatics tool, named the “VERA Revolution”, were developed to detect variants and CNVs in cfDNA samples. Results were compared with genomic DNA (gDNA) extracted from fetal samples, including amniotic fluid and chorionic villus sampling and buccal swabs. Results: The study included pregnant women with gestational ages from 10 + 3 to 15 + 2 weeks (mean: 12.1 weeks). The fetal fraction (FF), a crucial measure of cfDNA test reliability, ranged from 5% to 20%, ensuring adequate DNA amount for analysis. Among 36 families tested, 14 showed a wild-type genotype. Identified variants included two deletions (22q11.2, and 4p16.3), two duplications (16p13 and 5p15), and eighteen single-nucleotide variants (one in CFTR, three in GJB2, three in PAH, one in RIT1, one in DHCR7, one in TCOF1, one in ABCA4, one in MYBPC3, one in MCCC2, two in GBA1 and three in PTPN11). Significant concordance was found between our panel results and prenatal/postnatal genetic profiles. Conclusions: The “VERA Revolution” test highlights advancements in prenatal genomic screening, offering potential improvements in prenatal care.
Full article
(This article belongs to the Section Technologies and Resources for Genetics)
►▼
Show Figures
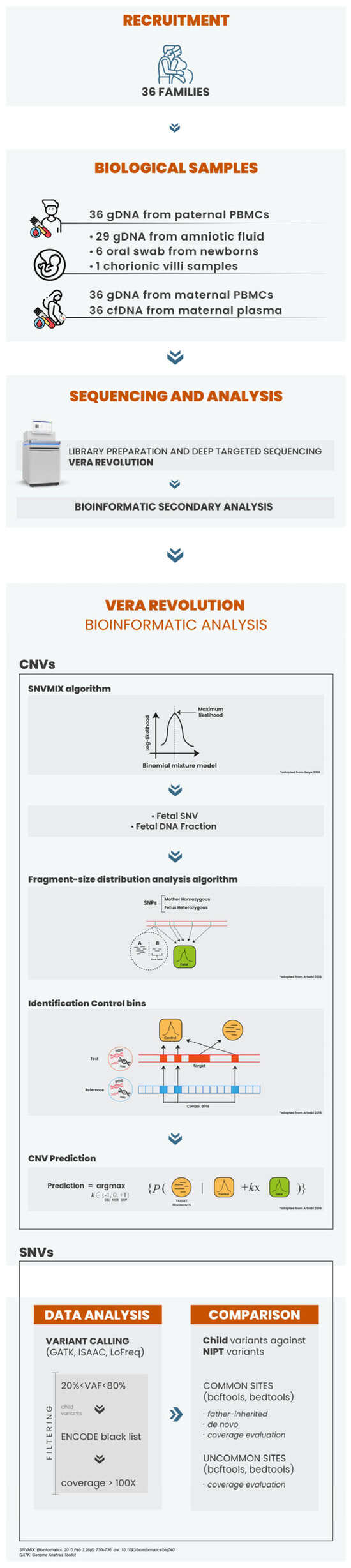
Figure 1
Open AccessReview
Tribe Paniceae Cereals with Different Ploidy Levels: Setaria italica, Panicum miliaceum, and Echinochloa esculenta
by
Kazuhiro Satomura
Genes 2025, 16(4), 426; https://doi.org/10.3390/genes16040426 - 1 Apr 2025
Abstract
Plants have repeatedly undergone whole-genome duplication during their evolutionary history. Even in modern plants, there is diversity in ploidy within and between species, providing a snapshot of the evolutionary turnover of ploidy. Here, I will review the diversity of ploidy and the evolution
[...] Read more.
Plants have repeatedly undergone whole-genome duplication during their evolutionary history. Even in modern plants, there is diversity in ploidy within and between species, providing a snapshot of the evolutionary turnover of ploidy. Here, I will review the diversity of ploidy and the evolution of the genome constitution, focusing on the millet species Setaria italica, Panicum miliaceum, and Echinochloa esculenta. These are all historically important cereal crops that have been domesticated in East Asia. They all display a basic chromosome set of nine, but they are diploid, tetraploid, and hexaploid, respectively. The timing of ploidy is different among the millet species, as is the extent of gene family expansion and gene loss. There also exists complex subgenomic evolution in the wild species within each genus. These three millet species and their related wild species are suitable models for elucidating the molecular evolution and diversity of genome duplication by comparative genomic analysis.
Full article
(This article belongs to the Special Issue Gene and Genome Duplications in Plants)
►▼
Show Figures

Figure 1
Open AccessArticle
GONNMDA: A Ordered Message Passing GNN Approach for miRNA–Disease Association Prediction
by
Sihao Zeng, Shanwen Zhang, Zhen Wang, Chen Yang and Shenao Yuan
Genes 2025, 16(4), 425; https://doi.org/10.3390/genes16040425 - 1 Apr 2025
Abstract
Small non-coding molecules known as microRNAs (miRNAs) play a critical role in disease diagnosis, treatment, and prognosis evaluation. Traditional wet-lab methods for validating miRNA–disease associations are often time-consuming and inefficient. With the advancement of high-throughput sequencing technologies, deep learning methods have become effective
[...] Read more.
Small non-coding molecules known as microRNAs (miRNAs) play a critical role in disease diagnosis, treatment, and prognosis evaluation. Traditional wet-lab methods for validating miRNA–disease associations are often time-consuming and inefficient. With the advancement of high-throughput sequencing technologies, deep learning methods have become effective tools for uncovering potential patterns in miRNA–disease associations and revealing novel biological insights. Most of the existing approaches focus primarily on individual molecular behavior, overlooking interactions at the multi-molecular level. Conventional graph neural network (GNN) models struggle to generalize to heterogeneous graphs, and as network depth increases, node representations become indistinguishable due to over-smoothing, resulting in reduced predictive performance. GONNMDA first integrates similarity features from multiple data sources and applies noise reduction to obtain a reconstructed, comprehensive similarity representation. It then constructs heterogeneous graphs and applies a root–tree hierarchical alignment, along with an ordered gating message-passing mechanism, effectively addressing the challenges of heterogeneity and over-smoothing. Finally, a multilayer perceptron is employed to produce the final association predictions. To evaluate the effectiveness of GONNMDA, we conducted extensive experiments where the model achieved an AUC of 95.49% and an AUPR of 95.32%. The results demonstrate that GONNMDA outperforms several recent state-of-the-art methods. In addition, case studies and survival analyses on three common human cancers—breast cancer, rectal cancer, and lung cancer—further validate the effectiveness and reliability of GONNMDA in predicting miRNA–disease associations.
Full article
(This article belongs to the Section Bioinformatics)
►▼
Show Figures

Figure 1
Open AccessArticle
LMFE: A Novel Method for Predicting Plant LncRNA Based on Multi-Feature Fusion and Ensemble Learning
by
Hongwei Zhang, Yan Shi, Yapeng Wang, Xu Yang, Kefeng Li, Sio-Kei Im and Yu Han
Genes 2025, 16(4), 424; https://doi.org/10.3390/genes16040424 - 31 Mar 2025
Abstract
Background/Objectives: Long non-coding RNAs (lncRNAs) play a crucial regulatory role in plant trait expression and disease management, making their accurate prediction a key research focus for guiding biological experiments. While extensive studies have been conducted on animals and humans, plant lncRNA research
[...] Read more.
Background/Objectives: Long non-coding RNAs (lncRNAs) play a crucial regulatory role in plant trait expression and disease management, making their accurate prediction a key research focus for guiding biological experiments. While extensive studies have been conducted on animals and humans, plant lncRNA research remains relatively limited due to various challenges, such as data scarcity and genomic complexity. This study aims to bridge this gap by developing an effective computational method for predicting plant lncRNAs, specifically by classifying transcribed RNA sequences as lncRNAs or mRNAs using multi-feature analysis. Methods: We propose the lncRNA multi-feature-fusion ensemble learning (LMFE) approach, a novel method that integrates 100-dimensional features from RNA biological properties-based, sequence-based, and structure-based features, employing the XGBoost ensemble learning algorithm for prediction. To address unbalanced datasets, we implemented the synthetic minority oversampling technique (SMOTE). LMFE was validated across benchmark datasets, cross-species datasets, unbalanced datasets, and independent datasets. Results: LMFE achieved an accuracy of 99.42%, an F1score of 0.99, and an MCC of 0.98 on the benchmark dataset, with robust cross-species performance (accuracy ranging from 89.30% to 99.81%). On unbalanced datasets, LMFE attained an average accuracy of 99.41%, representing a 12.29% improvement over traditional methods without SMOTE (average ACC of 87.12%). Compared to state-of-the-art methods, such as CPC2 and PLEKv2, LMFE consistently outperformed them across multiple metrics on independent datasets (with an accuracy ranging from 97.33% to 99.21%), with redundant features having minimal impact on performance. Conclusions: LMFE provides a highly accurate and generalizable solution for plant lncRNA prediction, outperforming existing methods through multi-feature fusion and ensemble learning while demonstrating robustness to redundant features. Despite its effectiveness, variations in performance across species highlight the necessity for future improvements in managing diverse plant genomes. This method represents a valuable tool for advancing plant lncRNA research and guiding biological experiments.
Full article
(This article belongs to the Section Bioinformatics)
►▼
Show Figures
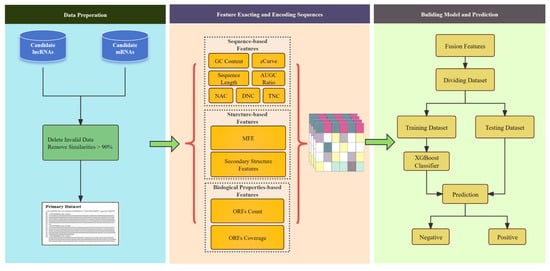
Figure 1
Open AccessReview
Ethical and Practical Considerations in Implementing Population-Based Reproductive Genetic Carrier Screening
by
Eva Van Steijvoort and Pascal Borry
Genes 2025, 16(4), 423; https://doi.org/10.3390/genes16040423 - 31 Mar 2025
Abstract
Reproductive genetic carrier screening (RGCS) has emerged as a promising tool for identifying couples with an increased likelihood of conceiving a child with an autosomal recessive or X-linked genetic condition. By enabling early detection, RGCS has the potential to support informed reproductive decision-making.
[...] Read more.
Reproductive genetic carrier screening (RGCS) has emerged as a promising tool for identifying couples with an increased likelihood of conceiving a child with an autosomal recessive or X-linked genetic condition. By enabling early detection, RGCS has the potential to support informed reproductive decision-making. Historically, carrier screening initiatives aimed to decrease the prevalence of specific genetic disorders by targeting particular high-risk populations. More recently, there has been a shift towards offering RGCS for a wider range of conditions, with the goal of enhancing reproductive autonomy by facilitating informed decision-making and addressing inequities in access to healthcare interventions. However, this shift towards a more inclusive, population-based approach has raised questions about the tension between individual autonomy and public health goals, as well as concerns regarding the potential negative effects of large-scale genetic screening initiatives. Furthermore, there is growing interest in utilizing RGCS data for broader purposes, such as population-based genetic screening programs for hereditary cancers or identifying causes of unexplained infertility, which may present additional ethical considerations. This review explores the complexities surrounding the implementation of RGCS, with an emphasis on its objectives, the significance of informed decision-making, and the wider societal challenges it may present. By analyzing these interconnected factors, we aim to provide a thorough understanding of the potential implications of RGCS on both individual autonomy and societal dynamics.
Full article
(This article belongs to the Special Issue Human Genetics: Diseases, Community, and Counseling)
Open AccessArticle
Genome-Wide Association Study Identifies Potential Regulatory Loci and Pathways Related to Buffalo Reproductive Traits
by
Wangchang Li, Qiyang Xie, Haiying Zheng, Anqin Duan, Liqing Huang, Chao Feng, Jianghua Shang and Chunyan Yang
Genes 2025, 16(4), 422; https://doi.org/10.3390/genes16040422 - 31 Mar 2025
Abstract
Background: The reproductive performance of water buffalo significantly impacts the economic aspects of production. Traditional breeding methods are constrained by low heritability and numerous influencing factors, making it difficult to effectively improve reproductive efficiency. Genome-wide association studies (GWAS) offer new possibilities for exploring
[...] Read more.
Background: The reproductive performance of water buffalo significantly impacts the economic aspects of production. Traditional breeding methods are constrained by low heritability and numerous influencing factors, making it difficult to effectively improve reproductive efficiency. Genome-wide association studies (GWAS) offer new possibilities for exploring reproductive traits in water buffalo, opening up new avenues for efficient breeding. Methods: Using whole-genome resequencing, we identified quantitative trait loci (QTLs) associated with four suggestive reproductive traits: calving interval (CI), calf birth weight (CBW), dam birth weight (BW), and age at first calving (FCA). The study focused on identifying genetic variants that influence these reproductive traits. Results: Our research identified 52 suggestive regulatory loci associated with reproductive traits in water buffalo. Based on a 50 kb interval, we annotated these loci to 58 candidate genes. These loci involve genes such as AGBL4, GRM1, NCKAP5, and NRXN1, which are primarily enriched in pathways including the FOXO signaling pathway, calcium ion pathways, estrogen signaling pathway, and phospholipase D signaling pathway. These pathways directly or indirectly regulate the reproductive efficiency of water buffalo. Conclusions: This study has revealed suggestive regulatory genes (AGBL4, GRM1, NCKAP5, NRXN1) associated with reproductive traits in water buffalo. This not only enhances our understanding of the molecular mechanisms underlying complex traits but also points towards strategies for improving the reproductive capacity of water buffalo. These findings provide a solid foundation for future breeding programs aimed at enhancing water buffalo productivity.
Full article
(This article belongs to the Special Issue Buffalo Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Identification of Novel Mosaic Variants in Focal Epilepsy-Associated Patients’ Brain Lesions
by
Camila Araújo Bernardino Garcia, Muhammad Zubair, Marcelo Volpon Santos, Sang Hyun Lee, Ian Alfred Graham, Valentina Stanley, Renee D. George, Joseph G. Gleeson, Hélio Rubens Machado and Xiaoxu Yang
Genes 2025, 16(4), 421; https://doi.org/10.3390/genes16040421 - 31 Mar 2025
Abstract
Focal cortical dysplasia type III (FCDIII) is a rare and complex condition associated with drug-resistant epilepsy and often characterized by cortical lamination abnormalities, along with a variety of neoplasms and vascular abnormalities. Objectives: This study aimed to elucidate the genetic architecture underlying FCDIII
[...] Read more.
Focal cortical dysplasia type III (FCDIII) is a rare and complex condition associated with drug-resistant epilepsy and often characterized by cortical lamination abnormalities, along with a variety of neoplasms and vascular abnormalities. Objectives: This study aimed to elucidate the genetic architecture underlying FCDIII through the use of whole-exome sequencing (WES) of brain and peripheral blood samples from 19 patients who had been diagnosed with FCDIII. Methods: Variants were identified through a series of machine-learning-based detection and functional prediction methods and were not previously associated with FCDIII. Mosaic fraction scores of these variants validated the variants’ pathogenicity, and in silico and gene ontology enrichment analyses demonstrated that these variants had severe destabilizing effects on protein structure. Results: We reported ten novel pathogenic somatic missense and loss of function variants across eight genes, including CNTNAP2, ACY1, SERAC1, and BRAF. Genetic alterations were linked to clinical manifestations, such as encephalopathies and intellectual disabilities, thereby emphasizing their role as molecular drivers of FCDIII. Conclusions: We demonstrated that next-generation sequencing-based mosaic variant-calling pipelines are useful for the genetic diagnosis of FCDIII, opening up avenues for targeted therapies, yet further research is required to validate these findings and examine their therapeutic implications.
Full article
(This article belongs to the Special Issue Genomic Mosaicism in Human Development and Diseases)
►▼
Show Figures

Figure 1
Open AccessReview
Aberrant Expression of Non-Coding RNAs in Pediatric T Acute Lymphoblastic Leukemia and Their Potential Application as Biomarkers
by
Neila Luciano, Luigi Coppola, Giuliana Salvatore, Pasquale Primo, Rosanna Parasole, Peppino Mirabelli and Francesca Maria Orlandella
Genes 2025, 16(4), 420; https://doi.org/10.3390/genes16040420 - 31 Mar 2025
Abstract
Less than 5% of the DNA sequence encodes for proteins, and the remainder encodes for non-coding RNAs (ncRNAs). Among the members of the ncRNA family, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) play a pivotal role in the insurgence and progression of several
[...] Read more.
Less than 5% of the DNA sequence encodes for proteins, and the remainder encodes for non-coding RNAs (ncRNAs). Among the members of the ncRNA family, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) play a pivotal role in the insurgence and progression of several cancers, including leukemia. Thought to have different molecular mechanisms, both miRNAs and lncRNAs act as epigenetic factors modulating gene expression and influencing hematopoietic differentiation, proliferation and immune system function. Here, we discuss the most recent findings on the main molecular mechanisms by which miRNAs and lncRNAs are involved in the pathogenesis and progression of pediatric T acute lymphoblastic leukemia (T-ALL), pointing out their potential utility as therapeutic targets and as biomarkers for early diagnosis, risk stratification and prognosis. miRNAs are involved in the pathogenesis of T-ALL, acting both as tumor suppressors and as oncomiRs. By contrast, to the best of our knowledge, the literature highlights lncRNAs as acting only as oncogenes in this type of cancer by inhibiting apoptosis and promoting cell cycle and drug resistance. Additionally, here, we discuss how these molecules could be detected in the plasma of T-ALL patients, highlighting that lncRNAs may represent a new class of promising accurate and sensitive biomarkers in these young patients. Thus, the unveiling of the aberrant signature of circulating and intracellular levels of lncRNAs could have great clinical utility for obtaining a more accurate definition of prognosis and uncovering novel therapeutic strategies against T-ALL in children. However, further investigations are needed to better define the standard methodological procedure for their quantification and to obtain their specific targeting in T-ALL pediatric patients.
Full article
(This article belongs to the Special Issue The Role of miRNAs in Human Cancer)
►▼
Show Figures

Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
2 April 2025
MDPI INSIGHTS: The CEO's Letter #21 - Annual Report, Swiss Consortium, IWD, ICARS, Serbia
MDPI INSIGHTS: The CEO's Letter #21 - Annual Report, Swiss Consortium, IWD, ICARS, Serbia
1 April 2025
Meet us at the 11th Congress of the European Academy of Neurology, 21–24 June 2025, Helsinki, Finland
Meet us at the 11th Congress of the European Academy of Neurology, 21–24 June 2025, Helsinki, Finland

Topics
Topic in
Biomolecules, Cells, Genes, ncRNA, IJMS
MicroRNA: Mechanisms of Action, Physio-Pathological Implications, and Disease Biomarkers, 3rd Edition
Topic Editors: Hsiuying Wang, Y-h. TaguchiDeadline: 30 April 2025
Topic in
Diversity, Forests, Genes, IJPB, Plants
Plant Chloroplast Genome and Evolution
Topic Editors: Chao Shi, Lassaâd Belbahri, Shuo WangDeadline: 31 August 2025
Topic in
Applied Sciences, BioMedInformatics, BioTech, Genes, Computation
Computational Intelligence and Bioinformatics (CIB)
Topic Editors: Marco Mesiti, Giorgio Valentini, Elena Casiraghi, Tiffany J. CallahanDeadline: 30 September 2025
Topic in
Agriculture, Agronomy, Crops, Genes, Plants, DNA
Vegetable Breeding, Genetics and Genomics, 2nd Volume
Topic Editors: Umesh K. Reddy, Padma Nimmakayala, Georgia NtatsiDeadline: 31 October 2025

Conferences
Special Issues
Special Issue in
Genes
Molecular Determinants and Pathophysiology of Eye Disorders
Guest Editor: Shi Song RongDeadline: 10 April 2025
Special Issue in
Genes
Molecular Breeding and Genetics of Plant Drought Resistance
Guest Editors: Ivan Pejić, Miroslav BukanDeadline: 10 April 2025
Special Issue in
Genes
Molecular Assays for Mutation and Infectious Agent Detection
Guest Editor: Gustavo BarraDeadline: 10 April 2025
Topical Collections
Topical Collection in
Genes
Study on Genotypes and Phenotypes of Pediatric Clinical Rare Diseases
Collection Editors: Livia Garavelli, Stefano Giuseppe Caraffi
Topical Collection in
Genes
Genotype-Phenotype Study in Disease
Collection Editors: Michele Cioffi, Maria Teresa Vietri
Topical Collection in
Genes
Feature Papers in ‘Animal Genetics and Genomics’
Collection Editors: Antonio Figueras, Raquel Vasconcelos






