-
 Relationship Between Human Microbiome and Helicobacter pylori
Relationship Between Human Microbiome and Helicobacter pylori -
 Sex Differences and Pathogen Patterns in Surgically Treated Aortic Valve Endocarditis over 15 Years
Sex Differences and Pathogen Patterns in Surgically Treated Aortic Valve Endocarditis over 15 Years -
 Transcriptomic Response of Balamuthia mandrillaris to Lippia graveolens Extract Fractions
Transcriptomic Response of Balamuthia mandrillaris to Lippia graveolens Extract Fractions -
 Broad Spectrum Antimicrobial Activity of Bacteria Isolated from Vitis vinifera Leaves
Broad Spectrum Antimicrobial Activity of Bacteria Isolated from Vitis vinifera Leaves
Journal Description
Microbiology Research
Microbiology Research
is an international, scientific, peer-reviewed open access journal published monthly online by MDPI (from Volume 11 Issue 2-2020).
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, ESCI (Web of Science), Embase, and other databases.
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.4 days after submission; acceptance to publication is undertaken in 2.7 days (median values for papers published in this journal in the second half of 2024).
- Recognition of Reviewers: APC discount vouchers, optional signed peer review, and reviewer names published annually in the journal.
Impact Factor:
2.1 (2023);
5-Year Impact Factor:
2.0 (2023)
Latest Articles
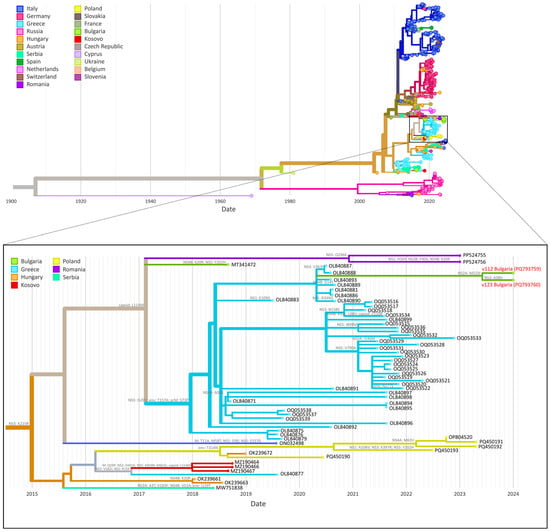
Epidemiological, Clinical and Phylogenetic Characteristics of West Nile Virus in Bulgaria, 2024
Microbiol. Res. 2025, 16(4), 82; https://doi.org/10.3390/microbiolres16040082 (registering DOI) - 4 Apr 2025
Abstract
►
Show Figures
West Nile Virus (WNV), a mosquito-borne pathogen, is a growing public health challenge across Europe. Environmental and anthropogenic factors have led to the spread of the virus to higher geographic latitudes, as well as to increased viral circulation and genetic diversity. Aims: This
[...] Read more.
West Nile Virus (WNV), a mosquito-borne pathogen, is a growing public health challenge across Europe. Environmental and anthropogenic factors have led to the spread of the virus to higher geographic latitudes, as well as to increased viral circulation and genetic diversity. Aims: This study aimed to describe the epidemiological, clinical, and laboratory characteristics of WNV cases in Bulgaria during 2024 and to investigate WNV phylogenetics. Epidemiological, clinical and laboratory data from 32 patients with confirmed or probable WNV infections were collected and analysed. Complete viral genomes were obtained from two samples using whole genome sequencing (WGS). Phylogenetic analysis was performed using the Nextstrain WNV analysis pipeline. Severe disease was observed in 21 patients, with three fatalities reported in older males with comorbidities. Phylogenetic analysis revealed that Bulgarian strains clustered within the Central/Southern European clade of lineage 2, closely related to Greek strains. Evidence suggested localised viral evolution following cross-border introduction from Greece. Our study provides a detailed clinical and laboratory characterization of the human WNV cases detected in Bulgaria in 2024. Improved diagnostic workflows, expanded laboratory resources and increased molecular surveillance are essential to better understand the burden of WNV infections in Bulgaria, as well as to follow the evolution and spread of the virus.
Full article
Open AccessArticle
Molecular and Epidemiological Analysis of Carbapenem-Resistant Klebsiella pneumoniae in a Greek Tertiary Hospital: A Retrospective Study
by
Alexandra Myari, Petros Bozidis, Efthalia Priavali, Eleni Kapsali, Vasilios Koulouras, Georgia Vrioni and Konstantina Gartzonika
Microbiol. Res. 2025, 16(4), 81; https://doi.org/10.3390/microbiolres16040081 - 4 Apr 2025
Abstract
►▼
Show Figures
Carbapenemase-producing Klebsiella pneumoniae is responsible for multiple serious infections with high mortality rates. K. pneumoniae carbapenemases (KPCs) are the most commonly isolated carbapenemases worldwide. To study the epidemiological and molecular characteristics of KPC-producing K. pneumoniae (KPC-KP), we conducted a retrospective study at the
[...] Read more.
Carbapenemase-producing Klebsiella pneumoniae is responsible for multiple serious infections with high mortality rates. K. pneumoniae carbapenemases (KPCs) are the most commonly isolated carbapenemases worldwide. To study the epidemiological and molecular characteristics of KPC-producing K. pneumoniae (KPC-KP), we conducted a retrospective study at the University General Hospital of Ioannina, Greece. A total of 177 K. pneumoniae clinical strains from the period 2014–2015 were confirmed as KPC producers by polymerase chain reaction (PCR) and were further examined for the presence of blaVIM, blaNDM, blaTEM, blaSHV, and blaCTX-M genes. Using the amplification refractory mutation system (ARMS) method, we identified the presence of the KPC-2 allele in 130 strains and the KPC-9 allele in 47. Strains from both allele groups belonged to the sequence type 258 (ST258). KPC-9 was responsible for a distinct outbreak, considered part of the broader KPC-2 outbreak. Molecular characterization of selected KPC-KP isolates from the period 2021–2022 revealed their continued presence in our hospital. Comparison of the antimicrobial susceptibility profiles of the two alleles showed a statistically significant increase in minimum inhibitory concentration (MIC) for ceftazidime (p = 0.03) and higher resistance to amikacin (p = 0.012) and colistin (p < 0.001) for KPC-9 compared to the KPC-2 allele. The two KPC alleles had similar mortality rates. This study demonstrates the heterogeneity of resistance genes in carbapenem-resistant K. pneumoniae (CR-KP) within a single-hospital setting and underscores the need for immediate containment measures.
Full article

Figure 1
Open AccessArticle
Enhancing Diagnostic Resilience: Evaluation of Extraction Platforms and IndiMag Pathogen Kits for Rapid Animal Disease Detection
by
Anne Vandenburg-Carroll, Douglas G. Marthaler and Ailam Lim
Microbiol. Res. 2025, 16(4), 80; https://doi.org/10.3390/microbiolres16040080 - 3 Apr 2025
Abstract
The United States is facing outbreaks of highly pathogenic avian influenza H5N1 in birds and dairy cattle, along with threats of African swine fever, classical swine fever, and foot-and-mouth disease. While the National Animal Health Laboratory Network (NAHLN) depends on high-throughput testing, the
[...] Read more.
The United States is facing outbreaks of highly pathogenic avian influenza H5N1 in birds and dairy cattle, along with threats of African swine fever, classical swine fever, and foot-and-mouth disease. While the National Animal Health Laboratory Network (NAHLN) depends on high-throughput testing, the KingFisher Duo Prime, IndiMag 48s, and IndiMag 2 are viable alternatives to aid in outbreak assessments. This study evaluates extraction platforms and the IndiMag Pathogen Kit for detecting the previous listed pathogens. Samples and reference materials were extracted using the MagMAX Viral RNA Isolation Kit, MagMAX CORE Nucleic Acid Purification Kit, and IndiMag Pathogen Kit. Real-time RT-PCR was performed following NAHLN protocols to assess analytical and diagnostic performance. Comparable limits of detection across extraction chemistries, instrumentation, and pathogens were demonstrated, with PCR efficiency ranging between 82.5% and 107.6%. The precision variability was low, with the coefficient of variation ranging from 0.16% to 1.76%. Diagnostic sensitivity and specificity were 100%, with a kappa coefficient of 1.0, indicating strong agreement between methods. These findings support the KingFisher Duo Prime, IndiMag 48s, IndiMag 2, and IndiMag Pathogen Kits as reliable options for NAHLN-approved testing, increasing equipment and reagent alternatives to enhance diagnostic resilience and improve response capabilities to emerging animal health threats.
Full article
(This article belongs to the Special Issue Veterinary Microbiology and Diagnostics)
Open AccessArticle
Bacterial Profile and Antimicrobial Resistance Pattern from Different Clinical Specimens at Uttara Adhunik Medical College Hospital, Dhaka
by
Mahfuza Nasrin, Fahmida Begum, Mohammad Julhas Sujan, Hridika Talukder Barua, Zakir Hossain Habib, S M Shahriar Rizvi, Aninda Rahman, Alina Shaw, Abul Hasnat, Soo Young Kwon, Rezina Karim, Md. Shah Alam, Noshin Nawal, Mohammad Moniruzzaman Bhuiyan, Ahmed Taha Aboushady, Adam Clark, John Stelling, Sanjay Gautam, Florian Marks and Nimesh Poudyal
Microbiol. Res. 2025, 16(4), 79; https://doi.org/10.3390/microbiolres16040079 - 2 Apr 2025
Abstract
►▼
Show Figures
Introduction: Antimicrobial resistance (AMR) is a critical global public health issue, leading to prolonged illness, increased morbidity and mortality, and rising healthcare costs. The effectiveness of antibiotics is diminishing due to the emergence of resistant bacterial strains. This study aimed to determine the
[...] Read more.
Introduction: Antimicrobial resistance (AMR) is a critical global public health issue, leading to prolonged illness, increased morbidity and mortality, and rising healthcare costs. The effectiveness of antibiotics is diminishing due to the emergence of resistant bacterial strains. This study aimed to determine the bacterial profile and AMR patterns of clinical isolates at Uttara Adhunik Medical College Hospital (UAMCH), Dhaka. Methods: A retrospective study was conducted at UAMCH from January 2017 to December 2019. A total of 32,187 clinical specimens (urine, blood, stool, wound swabs/pus, and sputum) were processed, of which 4232 yielded positive cultures. Bacterial identification followed standard bacteriological methods, and antibiotic susceptibility was assessed using the Kirby–Bauer disc diffusion method per CLSI guidelines. Data analysis was conducted using WHONET and QAAPT. Results: The highest proportion of positive cultures was from urine (47.5%), followed by blood (35.0%) and wound swabs/pus (10.1%). The most common isolates were Escherichia coli (37.2%), Salmonella typhi (25.7%), and Klebsiella sp. (11.5%). Gram-negative bacteria showed high resistance to commonly used antibiotics such as amoxicillin/clavulanic acid, cefixime, and ceftriaxone, while the resistance rates were lower for gentamicin, amikacin, and meropenem. However, Acinetobacter sp. exhibited alarming resistance to all tested antibiotics. Conclusions: This study highlights concerning resistance patterns among bacterial isolates, emphasizing the need for ongoing AMR surveillance to inform treatment strategies and improve patient care in Bangladesh.
Full article

Figure 1
Open AccessReview
Bacterial Foodborne Diseases in Central America and the Caribbean: A Systematic Review
by
Nicole Severino, Claudia Reyes, Yumeris Fernandez, Vasco Azevedo, Luis Enrique De Francisco, Rommel T. Ramos, Luis Orlando Maroto-Martín and Edian F. Franco
Microbiol. Res. 2025, 16(4), 78; https://doi.org/10.3390/microbiolres16040078 - 1 Apr 2025
Abstract
►▼
Show Figures
Foodborne diseases (FBDs) represent a significant public health concern, particularly in regions like Central America and the Caribbean (CAC), where surveillance gaps due to a lack of resources, knowledge, and technical abilities hinder control over outbreaks. This review investigates the bacterial pathogens responsible
[...] Read more.
Foodborne diseases (FBDs) represent a significant public health concern, particularly in regions like Central America and the Caribbean (CAC), where surveillance gaps due to a lack of resources, knowledge, and technical abilities hinder control over outbreaks. This review investigates the bacterial pathogens responsible for FBDs, their prevalence, management challenges, and prevention strategies. This systematic review followed PRISMA guidelines, focusing on bacterial FBDs in CAC from 2000 to 2024. PubMed and Google Scholar were used as primary databases, supported by other sources to identify relevant studies. Inclusion criteria encompassed studies focusing on bacterial pathogens, prevalence, risk factors, and surveillance practices. Out of the 509 studies initially identified, 35 met the inclusion criteria. The most prevalent pathogens were Salmonella spp., Escherichia coli, Campylobacter spp., and Aliarcobacter spp., with contamination often associated with poultry, eggs, and vegetables. Key challenges included inadequate surveillance systems, limited resources, and inconsistent reporting practices. A more significant investment in pathogen monitoring, documentation, and education, along with technologies like whole-genome sequencing (WGS), is crucial. Institutional and governmental funding is vital to improve surveillance and strengthen regional risk analysis.
Full article
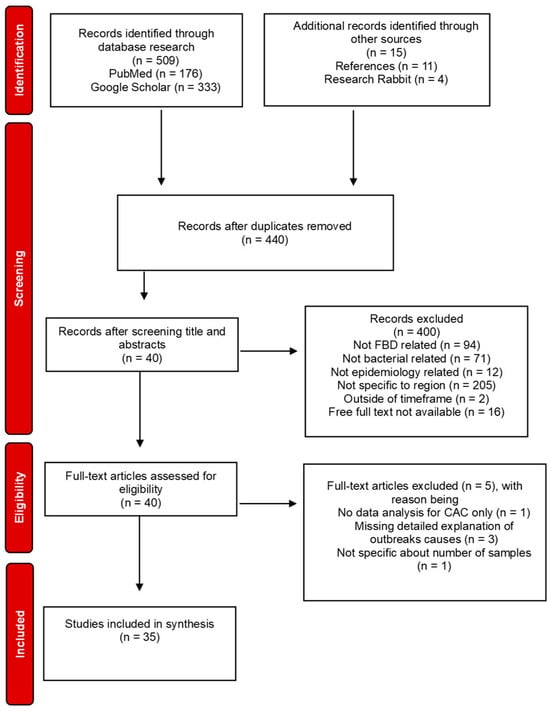
Figure 1
Open AccessArticle
Antiprotozoal Activity and Selectivity Index of Organic Salts of Albendazole and Mebendazole
by
Miriam Guadalupe Barón-Pichardo, Janeth Gómez-García, David Durán-Martínez, Oscar Torres-Angeles, Jesús Rivera-Islas and Blanca Estela Duque-Montaño
Microbiol. Res. 2025, 16(4), 77; https://doi.org/10.3390/microbiolres16040077 - 27 Mar 2025
Abstract
►▼
Show Figures
Infections from the protozoa Entamoeba histolytica (E. histolytica), Giardia lamblia (G. lamblia), and Trichomonas vaginalis (T. vaginalis) pose a public health issue, with albendazole and mebendazole serving as the second-line medications for treating these parasitic infections. However,
[...] Read more.
Infections from the protozoa Entamoeba histolytica (E. histolytica), Giardia lamblia (G. lamblia), and Trichomonas vaginalis (T. vaginalis) pose a public health issue, with albendazole and mebendazole serving as the second-line medications for treating these parasitic infections. However, the low aqueous solubility of these compounds has led to the exploration of new strategies to enhance their solubility, with the formation of salts being a commonly employed strategy. The sulfonates A1, A2, and A3 of albendazole, along with M1, M2, and M3 of mebendazole, were synthesized. The antiparasitic activity in vitro was assessed against the trophozoites of E. histolytica, G. lamblia, and T. vaginalis. The salts A2, A3, M2, and M3 demonstrated a greater antiparasitic effect (IC50 37.95–125.53 µM) compared to the positive controls albendazole and mebendazole. The salts A1, A3, M2, and M3 do not exhibit cytotoxic effects at concentrations of 500 µM on the Vero cell line. Taken together, these findings indicate that the formation of these new solid saline phases enhances the antiparasitic effects in vitro, which is crucial in the current search for improved, safe, and effective antiparasitic agents.
Full article
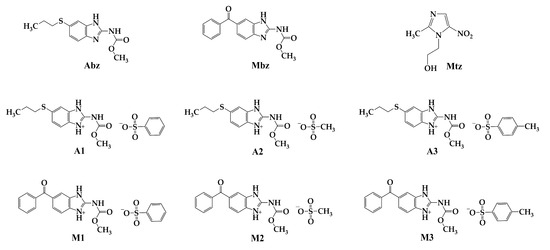
Figure 1
Open AccessReview
Rabbit Models for Infectious Diseases Caused by Staphylococcus aureus
by
Minghang Zeng, Yadong Wang, Fang Liu, Jinzhao Long and Haiyan Yang
Microbiol. Res. 2025, 16(4), 76; https://doi.org/10.3390/microbiolres16040076 - 27 Mar 2025
Abstract
►▼
Show Figures
Staphylococcus aureus (S. aureus) is a disreputable symbiotic bacterium that is responsible for a range of diseases, including life-threatening pneumonia, endocarditis, septicemia, and others, which has led to an immense loss in both public health and economy, imposing a significant burden
[...] Read more.
Staphylococcus aureus (S. aureus) is a disreputable symbiotic bacterium that is responsible for a range of diseases, including life-threatening pneumonia, endocarditis, septicemia, and others, which has led to an immense loss in both public health and economy, imposing a significant burden on society. To investigate the pathogenic mechanism of S. aureus and develop new treatment methods for infectious diseases caused by S. aureus, various rabbit models have been developed to simulate different infections by S. aureus, such as pneumonia models, meningitis models, and endocarditis models, etc. In this review, we summarized the application of rabbit models in S. aureus-induced infectious diseases.
Full article
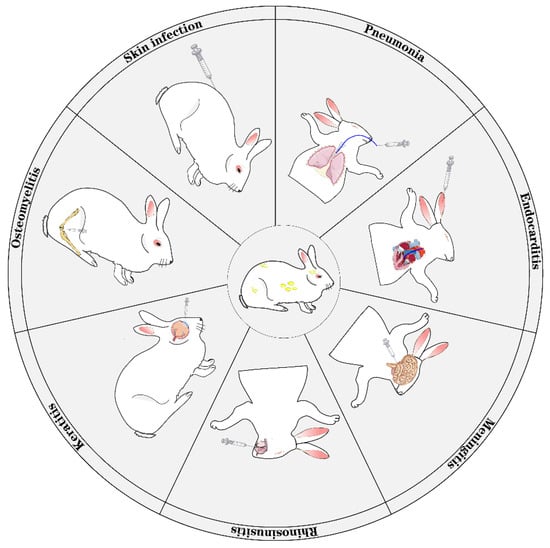
Figure 1
Open AccessReview
The Role of Microbial Dynamics, Sensorial Compounds, and Producing Regions in Cocoa Fermentation
by
Sofia de M. Campos, Walter J. Martínez-Burgos, Guilherme Anacleto dos Reis, Diego Yamir Ocán-Torres, Gabriela dos Santos Costa, Fernando Rosas Vega, Beatriz Alvarez Badel, Liliana Sotelo Coronado, Josilene Lima Serra and Carlos Ricardo Soccol
Microbiol. Res. 2025, 16(4), 75; https://doi.org/10.3390/microbiolres16040075 - 26 Mar 2025
Abstract
►▼
Show Figures
Cocoa fermentation is a critical step in chocolate production, influencing the flavor, aroma, and overall quality of the final product. This review focuses on the microbial dynamics of cocoa fermentation, emphasizing the roles of yeasts, lactic acid bacteria (LAB), and acetic acid bacteria
[...] Read more.
Cocoa fermentation is a critical step in chocolate production, influencing the flavor, aroma, and overall quality of the final product. This review focuses on the microbial dynamics of cocoa fermentation, emphasizing the roles of yeasts, lactic acid bacteria (LAB), and acetic acid bacteria (AAB). These microorganisms interact in a well-defined succession, producing organoleptic compounds such as alcohols, organic acids, and esters, which are key to the sensory profile of cocoa. This article examines the impact of different fermentation methods, including spontaneous fermentation and the use of starter cultures, on microbial communities and flavor development. Advances in starter culture technology are highlighted, demonstrating how microbial control can enhance fermentation efficiency, reduce fermentation time, and improve the consistency of chocolate flavor. Patents related to cocoa fermentation further emphasize the growing interest in microbial management to meet market demands for high-quality, distinct chocolate. This review also outlines future research directions, including the identification of new microbial strains, optimization of fermentation conditions, and the potential of biotechnological advancements to improve the fermentation process. Understanding microbial dynamics in cocoa fermentation offers significant potential for enhancing chocolate quality, sustainability, and the development of new, region-specific flavor profiles.
Full article
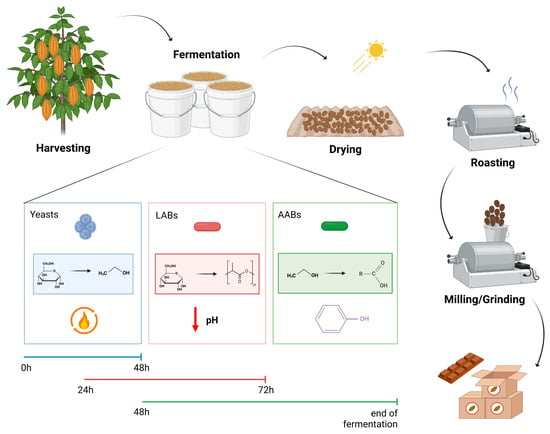
Figure 1
Open AccessArticle
Frequency, Resistance Patterns, and Serotypes of Salmonella Identified in Samples from Pigs of Colombia Collected from 2022 to 2023
by
Stefany Barrientos-Villegas, Juana L. Vidal, Nidia Gomez, Fernando L. Leite, Sara López-Osorio and Jenny J. Chaparro-Gutiérrez
Microbiol. Res. 2025, 16(4), 74; https://doi.org/10.3390/microbiolres16040074 - 25 Mar 2025
Abstract
►▼
Show Figures
The objective of this study was to determine the frequency of Salmonella in pig samples analyzed at the Veterinary Diagnostic Unit of the Faculty of Agricultural Sciences of the University of Antioquia, Colombia, between 2022 and 2023. Out of 5820 serum samples analyzed
[...] Read more.
The objective of this study was to determine the frequency of Salmonella in pig samples analyzed at the Veterinary Diagnostic Unit of the Faculty of Agricultural Sciences of the University of Antioquia, Colombia, between 2022 and 2023. Out of 5820 serum samples analyzed using indirect enzyme-linked immunosorbent assay, 63.76% were positive. Additionally, Salmonella was isolated and identified in a separate group of 848 samples (feces, tissues, etc.) with a positivity rate of 23.47%. Eight serotypes were identified, with the most common being Salmonella enterica subsp. enterica serotype Typhimurium (49.2%), followed by its monophasic variant I 4,[5],12:i:- (23%), and serotype Choleraesuis (18%). Antimicrobial susceptibility profiles of 105 isolates were evaluated using the Kirby–Bauer method, which demonstrated higher resistance (100%) to ampicillin, tiamulin, penicillin, tylosin, and erythromycin (these were the least tested), followed by florfenicol (44/54), doxycycline (4/5), spectinomycin (18/25), amoxicillin (32/46), chloramphenicol (2/3), tetracycline (2/3), and enrofloxacin (34/64), with lower resistance observed for fosfomycin (2/38) and ceftiofur (5/35). Multi-drug resistance was observed in 59% (62/105) of the isolates. The high proportion of Salmonella and the levels of resistance to various drugs raise significant concerns, indicating potential deficiencies in responsible antimicrobial use and management practices on pig farms in the region.
Full article
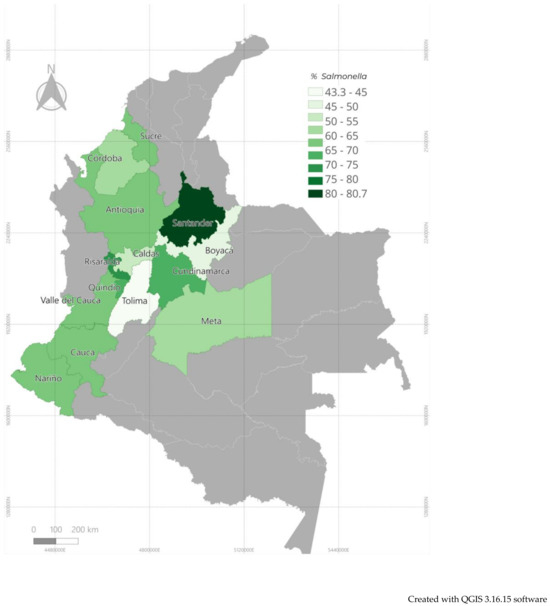
Figure 1
Open AccessReview
Molecular Epidemiology and Antimicrobial Resistance in Uropathogenic Escherichia coli in Saudi Arabian Healthcare Facilities
by
Mateq Ali Alreshidi
Microbiol. Res. 2025, 16(4), 73; https://doi.org/10.3390/microbiolres16040073 - 22 Mar 2025
Abstract
►▼
Show Figures
Urinary tract infections (UTIs) caused by uropathogenic Escherichia coli (UPEC) are a major healthcare challenge, necessitating effective antimicrobial therapy for treatment. However, the prevalence of antimicrobial resistance among UPEC strains is escalating, particularly among patients experiencing recurrent infection. The rise in UPEC strains
[...] Read more.
Urinary tract infections (UTIs) caused by uropathogenic Escherichia coli (UPEC) are a major healthcare challenge, necessitating effective antimicrobial therapy for treatment. However, the prevalence of antimicrobial resistance among UPEC strains is escalating, particularly among patients experiencing recurrent infection. The rise in UPEC strains that exhibit resistance to multiple antimicrobial agents, including the spread of extended-spectrum beta-lactamase (ESBL)-producing UPEC, intensifies the complexity of managing UTIs. Genetic variations within UPEC strains play a major role in their ability to resist antimicrobial agents and adapt to changing environments. Unveiling and understanding the genomic landscape of emerging UPEC strains is essential for comprehending the genetic basis of their resilience. Moreover, monitoring these genetic strains is crucial for identifying patterns of resistance dissemination, guiding infection control measures, and informing the development of targeted therapeutics.
Full article

Figure 1
Open AccessArticle
Inducing and Enhancing Antimicrobial Activity of Mining-Soil-Derived Actinomycetes Through Component Modification of Bennett’s Culture Medium
by
Soumia Ait Assou, Jaouad Anissi, Laurent Dufossé, Mireille Fouillaud and Mohammed EL Hassouni
Microbiol. Res. 2025, 16(4), 72; https://doi.org/10.3390/microbiolres16040072 - 22 Mar 2025
Abstract
►▼
Show Figures
This study investigated the effect of different culture agar media, derived from Bennett’s medium, on the antimicrobial activity of 15 Streptomyces sp. and 1 Lentzea sp. strains isolated from mining environments. The media were prepared from the standard Bennett’s medium by suppressing one,
[...] Read more.
This study investigated the effect of different culture agar media, derived from Bennett’s medium, on the antimicrobial activity of 15 Streptomyces sp. and 1 Lentzea sp. strains isolated from mining environments. The media were prepared from the standard Bennett’s medium by suppressing one, two, or three ingredients—yeast extract (YE), beef extract (BE), or casein (Cas)—while maintaining glucose (Gluc) or by substituting it with fructose (Fruc) or galactose (Gal) and keeping the same suppressions. The antimicrobial activity was investigated against Candida albicans ATCC 10231, Staphylococcus aureus ATCC 29213, Bacillus subtilis ATCC 6633, and Escherichia coli K12. The antimicrobial activity of actinomycete strains was positively influenced by media modifications, though the response was actinomycete strain and target pathogen-dependent. Unexpectedly, thirteen strains exhibited poor growth on a pure agar-agar medium, including six Streptomyces strains (AS34, AS3, BS59, BS68, BS69, and DAS104) that showed notable antimicrobial activity, with inhibition zone diameters ranging from 10.75 ± 1.06 to 18.00 ± 0.00 mm. Modifications of Bennett’s medium, including replacing glucose with fructose or galactose and maintaining yeast extract or both yeast extract and beef extract, induced and enhanced the antimicrobial activity of several actinomycete strains. Notably, the new media induced antimicrobial activity in strains that showed no activity in Bennett’s medium. They led, compared to Bennett’s medium, to the detection of eight additional active strains against S. aureus, eight against B. subtilis, six against E. coli, and four against C. albicans. This study is the first to explore the modification of Bennett’s medium, either by subtraction or substitution, in order to investigate the effect on antimicrobial activity of actinomycete strains. These results highlight the importance of the composition of culture media on inducing or boosting antimicrobial activity in Streptomyces and Lentzea.
Full article

Figure 1
Open AccessArticle
Shotgun Metagenomics Reveals Metabolic Potential and Functional Diversity of Microbial Communities of Chitu and Shala Soda Lakes in Ethiopia
by
Gessesse Kebede Bekele, Ebrahim M. Abda, Fassil Assefa Tuji, Abu Feyisa Meka and Mesfin Tafesse Gemeda
Microbiol. Res. 2025, 16(3), 71; https://doi.org/10.3390/microbiolres16030071 - 19 Mar 2025
Abstract
►▼
Show Figures
Soda lakes are extreme saline–alkaline environments that harbor metabolically versatile microbial communities with significant biotechnological potential. This study employed shotgun metagenomics (NovaSeq PE150) to investigate the functional diversity and metabolic potential of microbial communities in Ethiopia’s Chitu and Shala Lakes. An analysis of
[...] Read more.
Soda lakes are extreme saline–alkaline environments that harbor metabolically versatile microbial communities with significant biotechnological potential. This study employed shotgun metagenomics (NovaSeq PE150) to investigate the functional diversity and metabolic potential of microbial communities in Ethiopia’s Chitu and Shala Lakes. An analysis of gene content revealed 554,609 and 525,097 unique genes in Chitu and Shala, respectively, in addition to a substantial fraction (1,253,334 genes) shared between the two, underscoring significant functional overlap. Taxonomic analysis revealed a diverse phylogenetic composition, with bacteria (89% in Chitu Lake, 92% in Shala Lake) and archaea (4% in Chitu Lake, 0.8% in Shala Lake) as the dominant domains, alongside eukaryotes and viruses. Predominant bacterial phyla included Pseudomonadota, Actinomycetota, and Gemmatimonadota, while Euryarchaeota and Nitrososphaerota were prominent among archaea. Key genera identified in both lakes were Nitriliruptor, Halomonas, Wenzhouxiangella, Thioalkalivibrio, Aliidiomarina, Aquisalimonas, and Alkalicoccus. Functional annotation using the KEGG, eggNOG, and CAZy databases revealed that the identified unigenes were associated with various functions. Notably, genes related to amino acid, carbohydrate, and energy metabolism (KEGG levels 1–2) were predominant, indicating that conserved core metabolic functions are essential for microbial survival in extreme conditions. Higher-level pathways included quorum sensing, two-component signal transduction, and ABC transporters (KEGG level 3), facilitating environmental adaptation, stress response, and nutrient acquisition. The eggNOG annotation revealed that 13% of identified genes remain uncharacterized, representing a vast untapped reservoir of novel enzymes and biochemical pathways with potential applications in biofuels, bioremediation, and synthetic biology. This study identified 375 unique metabolic pathways, including those involved in pyruvate metabolism, xenobiotic degradation, lipid metabolism, and oxidative stress resistance, underscoring the microbial communities’ ability to thrive under fluctuating salinity and alkalinity. The presence of carbohydrate-active enzymes (CAZymes), such as glycoside hydrolases, polysaccharide lyases, and oxidoreductases, highlights their role in biomass degradation and carbon cycling. Enzymes such as alkaline proteases (Apr), lipases (Lip), and cellulases further support the lakes’ potential as sources of extremophilic biocatalysts. These findings position soda lakes as reservoirs of microbial innovation for extremophile biotechnology. Future research on unannotated genes and enzyme optimization promises sustainable solutions in bioenergy, agriculture, and environmental management.
Full article

Figure 1
Open AccessArticle
Antisense Oligonucleotide-Capped Gold Nanoparticles as a Potential Strategy for Tackling Antimicrobial Resistance
by
Cesar Rodolfo Garza-Cardenas, Angel Leon-Buitimea, A. A. Siller-Ceniceros and Jose Ruben Morones-Ramirez
Microbiol. Res. 2025, 16(3), 70; https://doi.org/10.3390/microbiolres16030070 - 18 Mar 2025
Abstract
►▼
Show Figures
Multidrug-resistant (MDR) bacterial pathogens pose a serious threat to global health, underscoring the urgent need for innovative therapeutic strategies. In this work, we designed and characterized thiol-modified antisense oligonucleotide-capped gold nanoparticles (ASO-AuNPs) to resensitize antibiotic-resistant bacteria. Transmission electron microscopy and UV–Vis spectroscopy confirmed
[...] Read more.
Multidrug-resistant (MDR) bacterial pathogens pose a serious threat to global health, underscoring the urgent need for innovative therapeutic strategies. In this work, we designed and characterized thiol-modified antisense oligonucleotide-capped gold nanoparticles (ASO-AuNPs) to resensitize antibiotic-resistant bacteria. Transmission electron microscopy and UV–Vis spectroscopy confirmed the morphology, size, and optical properties of AuNPs and ASO-AuNPs. Minimum inhibitory concentrations (MIC) of ampicillin were determined for non-resistant Escherichia coli DH5α (16 ppm) and an ampicillin-resistant E. coli DH5α strain (PSK, 32,768 ppm). When co-administered with ampicillin, ASO-AuNPs (0.1 and 0.2 nM) significantly reduced bacterial growth compared to the antibiotic-alone control (p < 0.05), demonstrating the capacity of ASO-AuNPs to restore antibiotic efficacy. These findings provide a proof of concept that antisense oligonucleotide-functionalized nanomaterials can be harnessed to overcome beta-lactam resistance, setting the stage for further optimization and translation into clinical applications.
Full article

Figure 1
Open AccessArticle
The Isolation of Free-Living Nitrogen-Fixing Bacteria and the Assessment of Their Potential to Enhance Plant Growth in Combination with a Commercial Biostimulant
by
Elodie Buisset, Martin Soust and Paul T. Scott
Microbiol. Res. 2025, 16(3), 69; https://doi.org/10.3390/microbiolres16030069 - 18 Mar 2025
Abstract
►▼
Show Figures
The development of microbial-based biostimulants to enhance the growth of crops and support a healthy and sustainable soil requires the isolation and large-scale industrial culture of effective microorganisms. In this study, work was undertaken to isolate and characterize free-living nitrogen-fixing bacteria capable of
[...] Read more.
The development of microbial-based biostimulants to enhance the growth of crops and support a healthy and sustainable soil requires the isolation and large-scale industrial culture of effective microorganisms. In this study, work was undertaken to isolate and characterize free-living nitrogen-fixing bacteria capable of acting as biostimulants alone or by incorporation into and/or supplementation with a current commercial crop biostimulant for farmers. Free-living bacteria were isolated from soil, sugar cane mulch, and plant roots following preliminary culture in a nitrogen-free media that targeted specific groups of known diazotrophs. Following the identification of each isolate by 16S rDNA sequence analysis, isolates selected for further study were identified as most closely related to Priestia megaterium, Sphingobium yanoikuyae, and Burkholderia paludis. Each isolate was investigated for its capacity to promote plant growth in nitrogen-free media. Wheat seedlings were inoculated with the isolates separately, together as a consortium, or in combination with the commercial biostimulant, Great Land Plus®. Compared to no-treatment control plants, the fresh weights were higher in both the shoots (183.2 mg vs. 330.6 mg; p < 0.05) and roots (320.4 mg vs. 731.3 mg; p < 0.05) of wheat seedlings inoculated with P. megaterium. The fresh weights were also higher in the shoots (267.8 mg; p < 0.05) and roots (610.3 mg; p = 0.05) of wheat seedlings inoculated with S. yanoikuyae. In contrast, the fresh weight of the shoot and root systems of plants inoculated with B. paludis were significantly lower (p < 0.05) than that of the no-treatment control plants. Moreover, when Great Land Plus® was supplemented with a consortium of P. megaterium and S. yanoikuyae, or a consortium of P. megaterium, S. yanoikuyae, and B. paludis no promotion of plant growth was observed.
Full article

Figure 1
Open AccessBrief Report
Effect of Tilapia Parvovirus (TiPV) on Fish Health: An In Vitro Approach
by
Vikash Kumar, Basanta Kumar Das, Anupam Adhikari, Kampan Bisai and Biswajit Mandal
Microbiol. Res. 2025, 16(3), 68; https://doi.org/10.3390/microbiolres16030068 - 17 Mar 2025
Abstract
Tilapia Parvovirus (TiPV) is a rising pathogen responsible for high mortality in tilapia aquaculture. Understanding TiPV’s pathogenesis is crucial for developing effective management strategies. This study aimed to elucidate TiPV pathogenesis by evaluating its cytotoxic effects on Danio rerio gill (DRG) cell monolayers
[...] Read more.
Tilapia Parvovirus (TiPV) is a rising pathogen responsible for high mortality in tilapia aquaculture. Understanding TiPV’s pathogenesis is crucial for developing effective management strategies. This study aimed to elucidate TiPV pathogenesis by evaluating its cytotoxic effects on Danio rerio gill (DRG) cell monolayers and its impact on host immune responses. PCR-confirmed TiPV-infected DRG cell monolayers were subjected to an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay at 24, 48, 72, and 96 h post-infection to assess cell viability and cytotoxicity. The MTT assay revealed a progressive decline in DRG cell viability over time, with viable cell percentages decreasing from 66.71% at 24 h to 31.28% at 96 h in TiPV-infected cultures, compared to consistently high viability in controls. Simultaneously, quantitative real-time PCR (qPCR) was used to assess the expression of key immune-related genes, including Interleukins (IL-1β, IL-8), Toll-like receptor 7 (TLR7), Major Histocompatibility Complex II (MHC-II), Tumor Necrosis Factor α (TNF-α), Nuclear Factor Kappa B (NF-κB), and Chemokine Receptors (CRs).qPCR analysis showed an upregulation of IL-8, IL-1β, TNF-α, and CRs, indicating an early inflammatory response. However, significant downregulation of TLR7, MHC-II, and NF-κB suggests TiPV’s ability to modulate host immune responses. The results highlight that TiPV induces significant cytotoxicity in DRG cells, leading to severe cellular damage. The virus also alters host immune responses by modulating the expression of key immune genes, which may contribute to its virulence and persistence. These findings enhance our understanding of TiPV pathogenesis and highlight the need for targeted research to develop effective control strategies for TiPV in aquaculture systems.
Full article
(This article belongs to the Special Issue Veterinary Microbiology and Diagnostics)
►▼
Show Figures

Figure 1
Open AccessReview
Microbial Metallophores in the Productivity of Agroecosystems
by
Lily X. Zelaya-Molina, Ismael F. Chávez-Díaz, José A. Urrieta-Velázquez, Marco A. Aragón-Magadan, Cristo O. Puente-Valenzuela, Mario Blanco-Camarillo, Sergio de los Santos-Villalobos and Juan Ramos-Garza
Microbiol. Res. 2025, 16(3), 67; https://doi.org/10.3390/microbiolres16030067 - 14 Mar 2025
Abstract
►▼
Show Figures
Microbial metallophores are low-molecular-weight chelating agents produced by microorganisms to acquire essential metal ions. Their biosynthesis, transport, and regulation involve complex processes, specialized enzymatic machinery, and intricate regulatory networks. This review examines the multifaceted roles of metallophores in microbial ecology and their potential
[...] Read more.
Microbial metallophores are low-molecular-weight chelating agents produced by microorganisms to acquire essential metal ions. Their biosynthesis, transport, and regulation involve complex processes, specialized enzymatic machinery, and intricate regulatory networks. This review examines the multifaceted roles of metallophores in microbial ecology and their potential applications in sustainable agriculture, emphasizing their key role in trace metal acquisition, nutrient cycling, and plant–microbe interactions. Furthermore, it explores the potential applications of metallophores in agriculture, bioremediation, and biotechnology, connecting their potential to the development of novel strategies for sustainable agriculture.
Full article

Figure 1
Open AccessSystematic Review
Prevention of Recurrent Urinary Tract Infection in Women: An Update
by
Elizabeth Corrales-Acosta, Eulalia Cuartiella Zaragoza, Mar Monzó Pérez, Sheila Benítez Perdomo, Juan Guillermo Corrales-Riveros and Mariela Corrales
Microbiol. Res. 2025, 16(3), 66; https://doi.org/10.3390/microbiolres16030066 - 11 Mar 2025
Abstract
►▼
Show Figures
Recurrent urinary tract infection (rUTI) is a significant public health problem in women. General measures to prevent recurrence include behavioral changes and increased fluid intake, cranberry ingest, use of methenamine hippurate, antibiotic prophylaxis, D-mannose, probiotics, or vaccines. We conducted a literature review of
[...] Read more.
Recurrent urinary tract infection (rUTI) is a significant public health problem in women. General measures to prevent recurrence include behavioral changes and increased fluid intake, cranberry ingest, use of methenamine hippurate, antibiotic prophylaxis, D-mannose, probiotics, or vaccines. We conducted a literature review of the latest updates on preventing rUTI in December 2024. The search concluded with 27 articles that fulfilled our inclusion criteria. Our review demonstrated that behavioral changes such as correct genital hygiene, avoiding postponing micturition or defecation, urinating after sexual intercourse, and ingesting 1.5–2 L of water could prevent rUTI. The ingestion of cranberries reduces the risk of symptomatic, culture-verified urinary tract infections in women with rUTIs. Methenamine hippurate is an alternative to antibiotics to avoid rUTI. Estrogen reduces rUTI in women with hypoestrogenism. Limited evidence supports using D-mannose, probiotics, and vaccines to prevent rUTI. In conclusion, after successful treatment of the acute episode, preventative measures are needed to reduce rUTI frequency and morbidity according to each patient’s characteristics and preferences.
Full article

Figure 1
Open AccessArticle
Using the bca Gene Coupled with a Tetracycline and Macrolide Susceptibility Profile to Identify the Highly Virulent ST283 Streptococcus agalactiae Strains in Thailand
by
Kwanchai Onruang, Panan Rattawongjirakul, Pisut Pongchaikul and Pitak Santanirand
Microbiol. Res. 2025, 16(3), 65; https://doi.org/10.3390/microbiolres16030065 - 10 Mar 2025
Abstract
Invasive infection by Streptococcus agalactiae (GBS) is a significant cause of death in newborn babies. In Thailand, data on strain distribution in GBS, specific virulence genes, and susceptibility patterns are limited. Therefore, our study aimed to establish the sequence type (ST) distribution and
[...] Read more.
Invasive infection by Streptococcus agalactiae (GBS) is a significant cause of death in newborn babies. In Thailand, data on strain distribution in GBS, specific virulence genes, and susceptibility patterns are limited. Therefore, our study aimed to establish the sequence type (ST) distribution and to use a specific virulence gene in combination with a susceptibility profile for strain identification. Non-duplicate 277 isolates of GBS were tested for ST, virulence genes, and antimicrobial susceptibility profiles. Twenty-five STs were detected. The ST283 (29.24%) and ST1 (27.07%) were the most common STs. The absence of the bca gene was an excellent marker to rule out ST283. All isolates were susceptible to nearly all tested antibiotics; however, only ST283 revealed 100% susceptibility to tetracycline, while ST1 and other non-ST283 showed 21.33 and 4.96%, respectively. Therefore, combining the alpha-C protein (bca) positive and tetracycline susceptible revealed 100% sensitivity for ST283. However, to identify the ST283, this combination revealed 78.9% specificity, which increased to 80.2% when erythromycin or azithromycin-susceptible was added. The bca positive combined with tetracycline and erythromycin susceptibility results were a simple tool for predicting ST283. The bca negative profile with tetracycline and macrolides resistance was commonly non-ST283. The information gained by this tool would benefit patient management.
Full article
Open AccessArticle
Microbiome of the Soil and Rhizosphere of the Halophyte Spergularia marina (L.) Griseb in the Saline Sites of Lake Kurgi, the South Urals: Metagenomic Analysis
by
Anastasia S. Tugbaeva, Alexander A. Ermoshin, Gregory I. Shiryaev and Irina S. Kiseleva
Microbiol. Res. 2025, 16(3), 64; https://doi.org/10.3390/microbiolres16030064 - 10 Mar 2025
Abstract
►▼
Show Figures
The study of the metagenomes of bacterial communities in saline areas is relevant in connection with the global salinization of agricultural lands. The aim of this study was to investigate the biodiversity and structure of rhizobacterial communities associated with the halophyte S. marina
[...] Read more.
The study of the metagenomes of bacterial communities in saline areas is relevant in connection with the global salinization of agricultural lands. The aim of this study was to investigate the biodiversity and structure of rhizobacterial communities associated with the halophyte S. marina from low and moderate sulfate–chloride salinity habitats. The bacterial community of bulk and rhizosphere soil was analyzed using high-throughput sequencing of the V1–V9 region of 16S rRNA by Oxford Nanopore Technologies. Alpha and beta diversity indices were calculated. A total of 55 phyla and 309 genera of bacteria were identified, among which Proteobacteria and Bacteroidetes dominated. The occurrence of Planctomycetes, Verrucomicrobia, and Acidobacteria in the rhizosphere was higher than in the bulk soil. Bacterial alpha diversity in the bulk soil decreased with increasing salinity, while it increased in the rhizosphere. The proportion of the halotolerant bacteria of Flavobacterium and Alteromonas genera significantly grew with increasing salinity both in the bulk and rhizosphere soil. In addition, in the rhizosphere, the percentage of Comamonas, Methylibium, Lysobacter, Planctomyces, Sphingomonas, Stenotrophomonas, and Lewinella genera increased. Among them, several genera included plant growth promoting rhizobacteria (PGPR). In the more saline bulk soil, the proportion of halotolerant genera Bacillus, Salinimicrobium, Marinobacter, Clostridium, Euzebya, KSA1, Marinobacter, Clostridium, Salinimicrobium, and Halorhodospira was also higher compared to the low saline site. Thus, increasing the salinity changed the taxonomic structure of the bacterial communities of both bulk soil and rhizosphere.
Full article

Figure 1
Open AccessArticle
Antimicrobial Activity and the Synergy Potential of Cinnamomum aromaticum Nees and Syzygium aromaticum (L.) Merr. et Perry Essential Oils with Antimicrobial Drugs
by
Soraia El Baz, Bouchra Soulaimani, Imane Abbad, Zineb Azgaou, El Mostapha Lotfi, Mustapha Malha and Noureddine Mezrioui
Microbiol. Res. 2025, 16(3), 63; https://doi.org/10.3390/microbiolres16030063 - 10 Mar 2025
Abstract
Antimicrobial resistance is a growing global challenge, rendering many standard treatments ineffective. Essential oils (EOs) of cinnamon (Cinnamomum aromaticum Nees) and clove (Syzygium aromaticum (L.) Merr. et Perry) may offer an alternative solution due to their high antimicrobial properties and their
[...] Read more.
Antimicrobial resistance is a growing global challenge, rendering many standard treatments ineffective. Essential oils (EOs) of cinnamon (Cinnamomum aromaticum Nees) and clove (Syzygium aromaticum (L.) Merr. et Perry) may offer an alternative solution due to their high antimicrobial properties and their abilities to fight resistant pathogens. This study evaluates the antimicrobial activity of these two EOs, and their synergistic potential when combined with two antibiotics (ciprofloxacin and vancomycin) and two antifungals (fluconazole and amphotericin B) against various bacterial and yeasts strains. The antimicrobial activities of each EO were evaluated by agar diffusion and broth microdilution assays, while the synergetic effects with antimicrobials were determined by calculating the fractional inhibitory concentration index (FICI) using the checkerboard method. The chemical composition of the EOs was analyzed using Gas Chromatography-Mass Spectrometry (GC-MS). The identification of individual components in the EOs was achieved by comparing their mass spectra with the NIST MS Search database and by correlating their retention times with those of known standards. GC-MS analysis revealed that the main constituents of S. aromaticum EO were eugenol (71.49%) and β-caryophyllene (23.43%), while C. aromaticum EO were dominated by cinnamaldehyde (47,04%) and cinnamyl acetate (18.93%). Antimicrobial activity showed that cinnamon EO exhibits highest effectiveness against all tested strains, with inhibition zones (IZ) ranging from 16.99 mm to 53.16 mm, and minimum inhibitory concentrations (MIC) and minimum microbicidal concentrations (MMC) ranging from 0.039 mg/mL to 0.156 mg/mL. However, for clove EO, the IZ ranged from 9.31 mm to 29.91 mm, with MIC and MMC values from 0.313 mg/mL to 1.25 mg/mL. In combination with antibiotics (ciprofloxacin and vancomycin), the studied EOs showed promising synergistic effects with reduction up to 128-fold. As regards antifungals (amphotericin B, and fluconazole), the synergistic effects were recorded with MIC gains up to 32-fold. Our findings demonstrate that the EOs from C. aromaticum and S. aromaticum exhibit significant broad-spectrum antimicrobial activity against diverse yeast and bacterial strains. This highlights their potential as bases for the development of novel plant-based antimicrobial agents. Importantly, the observed synergistic effects of these EOs with conventional antibiotics support their integration into medical treatments as a strategy to address microbial resistance. Future research should aim to elucidate the mechanisms underlying these synergistic actions, optimize their application, and enhance their therapeutic efficacy.
Full article
Highly Accessed Articles
Latest Books
E-Mail Alert
News
2 April 2025
MDPI INSIGHTS: The CEO's Letter #21 - Annual Report, Swiss Consortium, IWD, ICARS, Serbia
MDPI INSIGHTS: The CEO's Letter #21 - Annual Report, Swiss Consortium, IWD, ICARS, Serbia
24 March 2025
World Tuberculosis Day 2025—“Yes! We Can End TB: Commit, Invest, Deliver”, 24 March 2025
World Tuberculosis Day 2025—“Yes! We Can End TB: Commit, Invest, Deliver”, 24 March 2025

Topics
Topic in
Antibiotics, Antioxidants, JoF, Microbiology Research, Microorganisms
Redox in Microorganisms, 2nd Edition
Topic Editors: Michal Letek, Volker BehrendsDeadline: 31 July 2025
Topic in
JoF, Microbiology Research, Microorganisms, Pathogens
Pathophysiology and Clinical Management of Fungal Infections
Topic Editors: Allan J. Guimarães, Marcos de Abreu AlmeidaDeadline: 30 November 2025
Topic in
Applied Microbiology, Fermentation, Foods, Microbiology Research, Microorganisms
Fermented Food: Health and Benefit
Topic Editors: Niel Van Wyk, Alice VilelaDeadline: 31 December 2025
Topic in
Applied Microbiology, Microbiology Research, Microorganisms, IJMS, IJPB
New Challenges on Plant–Microbe Interactions
Topic Editors: Wenfeng Chen, Junjie ZhangDeadline: 31 January 2026

Conferences
Special Issues
Special Issue in
Microbiology Research
Antileishmanial Agents
Guest Editor: Edson Roberto da SilvaDeadline: 31 August 2025
Special Issue in
Microbiology Research
Probiotics, Pebiotics and Pet Health
Guest Editors: Bing Han, Lihong ZhaoDeadline: 30 September 2025
Topical Collections
Topical Collection in
Microbiology Research
Microbiology and Technology of Fermented Foods
Collection Editor: Salam A. Ibrahim
Topical Collection in
Microbiology Research
Public Health and Quality Aspects Related to Animal Productions
Collection Editors: Beniamino T. Cenci-Goga, Massimo Zerani, Luca Grispoldi
Topical Collection in
Microbiology Research
Microorganisms and Their Incredible Potential to Face Societal Challenges
Collection Editor: Mireille Fouillaud