-
 The Spleen Virome of Australia’s Endemic Platypus Is Dominated by Highly Diverse Papillomaviruses
The Spleen Virome of Australia’s Endemic Platypus Is Dominated by Highly Diverse Papillomaviruses -
 Marek’s Disease Virus (MDV) Meq Oncoprotein Plays Distinct Roles in Tumor Incidence, Distribution, and Size
Marek’s Disease Virus (MDV) Meq Oncoprotein Plays Distinct Roles in Tumor Incidence, Distribution, and Size -
 Fluorescent Clade IIb Lineage B.1 Mpox Viruses for Antiviral Screening
Fluorescent Clade IIb Lineage B.1 Mpox Viruses for Antiviral Screening -
 Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease
Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease -
 The Dissemination of Rift Valley Fever Virus to the Eye and Sensory Neurons of Zebrafish Larvae Is Stat1-Dependent
The Dissemination of Rift Valley Fever Virus to the Eye and Sensory Neurons of Zebrafish Larvae Is Stat1-Dependent
Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, AGRIS, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 17.1 days after submission; acceptance to publication is undertaken in 2.7 days (median values for papers published in this journal in the second half of 2024).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journal: Zoonotic Diseases.
Impact Factor:
3.8 (2023);
5-Year Impact Factor:
4.0 (2023)
Latest Articles
Cytomegalovirus Seroprevalence in Northern Poland in the Population Planning Pregnancy and Pregnant Women
Viruses 2025, 17(4), 537; https://doi.org/10.3390/v17040537 (registering DOI) - 7 Apr 2025
Abstract
Cytomegalovirus is an enveloped DNA virus. All forms of CMV infection—primary infection, reactivation, and infection with a different strain—may be asymptomatic. The risk of vertical transmission in the periconceptional period is approximately 20%, the risk of primary infection in the first trimester is
[...] Read more.
Cytomegalovirus is an enveloped DNA virus. All forms of CMV infection—primary infection, reactivation, and infection with a different strain—may be asymptomatic. The risk of vertical transmission in the periconceptional period is approximately 20%, the risk of primary infection in the first trimester is approximately 30%, and in the third trimester the risk increases to 70%. However, the most severe forms of congenital cytomegaly in newborns are related to infections in the periconceptional period. Offering a vaccine to the seronegative patients planning pregnancy may decrease incidents of congenital cytomegaly in neonates. The authors performed retrospective analysis of seroprevalence of CMV in 909 women who reported for pre-conceptional visits or routine pregnancy follow-ups (2003–2023). In the analyzed group, 577 (63.7%) women were seropositive. No influence related to the women’s age and place of residence was found. Higher seroprevalence was observed in women with children or those working in contact with many people. In the group of 332 seronegative patients, 21 (0.6%) were diagnosed with primary infection during pregnancy. Vaccinating 36.3% of patients planning pregnancy could significantly decrease the risk of primary infection during pregnancy, vertical transmission of CMV, and symptomatic infection in the neonates.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
Open AccessArticle
The Epidemiology of Hepatitis E in Israel and Potential Risk Factors: A Cross-Sectional Population-Based Serological Survey of Hepatitis E Virus in Northern Israel
by
Rasha Daniel, Shira Zelber-Sagi, Mira Barak and Eli Zuckerman
Viruses 2025, 17(4), 536; https://doi.org/10.3390/v17040536 (registering DOI) - 7 Apr 2025
Abstract
Hepatitis E Virus (HEV) has gained public health attention as one of the causative agents of viral hepatitis. Our study aimed to provide data about HEV seropositivity in the Israeli general population, including its seroprevalence geographical distribution, and to identify variables as possible
[...] Read more.
Hepatitis E Virus (HEV) has gained public health attention as one of the causative agents of viral hepatitis. Our study aimed to provide data about HEV seropositivity in the Israeli general population, including its seroprevalence geographical distribution, and to identify variables as possible risk factors for HEV exposure. A seroprevalence cross-sectional study was conducted: HEV serological status was determined in 716 blood samples collected from the routine check-up blood samples. Demographic information was available for all samples. The overall prevalence of HEV IgG in an apparently healthy population in the north of Israel was 10.5%, with no evidence of positive HEV IgM. There was a significant association between HEV seropositivity and elderly age and low socioeconomic status (SES). The age-adjusted seroprevalence was significantly lower among Jews compared to Arabs with a rate ratio of 2.02. We identified clusters (hot spots) of HEV infection in three regions under study. Our results confirmed a high prevalence of anti-HEV in the country where clinical hepatitis E is not endemic. For the first time, this study showed that a hot spot analysis was able to provide new knowledge about actual exposure zones. As HEV infection is not a notifiable disease, it is probably underdiagnosed. Thus, better awareness among physicians is warranted.
Full article
(This article belongs to the Special Issue Hepatitis E: Molecular Virology, Pathogenesis, and Treatment, 2nd Edition)
►▼
Show Figures
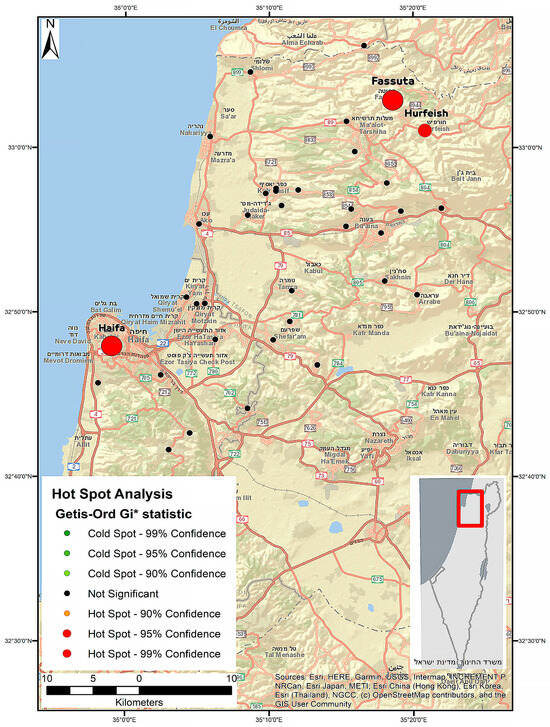
Figure 1
Open AccessReview
Landscape of H5 Infections in ASEAN Region: Past Insights, Present Realities, & Future Strategies
by
Muhammad Nur Adam Hatta, Yi Xin Nga, Ezryn Najwa Amirnuddin, Siti Nuraisyah Muzafar and Jasmine Elanie Khairat
Viruses 2025, 17(4), 535; https://doi.org/10.3390/v17040535 (registering DOI) - 6 Apr 2025
Abstract
The H5 Avian Influenza A virus infection has emerged as a global concern, particularly in the ASEAN region. This viral infection poses a significant threat to the poultry industry, public health, and regional economies. This region’s reliance on poultry production and the zoonotic
[...] Read more.
The H5 Avian Influenza A virus infection has emerged as a global concern, particularly in the ASEAN region. This viral infection poses a significant threat to the poultry industry, public health, and regional economies. This region’s reliance on poultry production and the zoonotic potential of H5 subtypes, with documented transmission to various mammalian species and humans, necessitates proactive mitigation strategies. Over the years, comprehensive efforts such as surveillance, vaccination programs, biosecurity measures, and public health education have been implemented to keep outbreaks at bay. In this review, we provide a thorough overview of the H5 infections in the ASEAN region, focusing on the unique challenges and successes in this geographic area. We analyze epidemiological trends, including specific high-risk populations and transmission patterns, and assess the socioeconomic impact of H5 outbreaks on local communities. We also examine regional responses, highlighting innovative surveillance programs, vaccination strategies, and biosecurity measures implemented to control the virus. Furthermore, we explore the crucial role of the One Health approach, emphasizing interdisciplinary collaboration between human, animal, and environmental health sectors. Finally, we discuss future strategies for prevention and control, including the importance of regional cooperation in combating this evolving threat. Through this, we aim to provide valuable insights to the public, policymakers, and researchers involved in tackling H5 infections globally.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
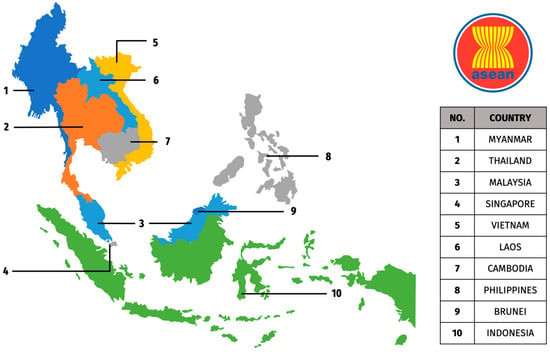
Figure 1
Open AccessArticle
Genomic and Epidemiological Investigations Reveal Chromosomal Integration of the Acipenserid Herpesvirus 3 Genome in Lake Sturgeon Acipenser fulvescens
by
Sharon Clouthier, Umberto Rosani, Arfa Khan, Qiuwen Ding, Eveline Emmenegger, Zhuozhi Wang, Thomas Nalpathamkalam and Bhooma Thiruvahindrapuram
Viruses 2025, 17(4), 534; https://doi.org/10.3390/v17040534 (registering DOI) - 5 Apr 2025
Abstract
DNA sequence from a new alloherpesvirus named acipenserid herpesvirus 3 (AciHV-3) was found in sturgeon species that are vulnerable to decline globally. A study was undertaken to develop a better understanding of the virus genome and to develop diagnostic tools to support an
[...] Read more.
DNA sequence from a new alloherpesvirus named acipenserid herpesvirus 3 (AciHV-3) was found in sturgeon species that are vulnerable to decline globally. A study was undertaken to develop a better understanding of the virus genome and to develop diagnostic tools to support an epidemiological investigation. A 184,426 bp genome was assembled from PacBio HiFi sequences generated with DNA from a Lake Sturgeon Acipenser fulvescens gonad cell line. The AciHV-3 genome was contiguous with host chromosomal DNA and was structured with telomere-like terminal direct repeat regions, five internal direct repeat regions and a U region that included intact open reading frames encoding alloherpesvirus core proteins. Diagnostic testing conducted with a newly developed and analytically validated qPCR assay established the ubiquitous presence and high titer of AciHV-3 DNA in somatic and germline tissues from wild Lake Sturgeon in the Hudson Bay drainage basin. Phylogenetic reconstructions confirm that the monophyletic AciHV-3 lineage shares a common ancestor with AciHV-1 and that AciHV-3 taxa cluster according to their sturgeon host. The same genotype of AciHV-3 is found in disjunctive Lake Sturgeon populations within and among drainage basins. The results support the hypotheses that AciHV-3 has established latency through germline chromosomal integration, is vertically transmitted via a Mendelian pattern of inheritance, is evolving in a manner consistent with a replication competent virus and has co-evolved with its host reaching genetic fixation in Lake Sturgeon populations in central Canada.
Full article
(This article belongs to the Special Issue Animal Herpesvirus)
►▼
Show Figures

Figure 1
Open AccessArticle
Phylodynamic of Tomato Brown Rugose Fruit Virus and Tomato Chlorosis Virus, Two Emergent Viruses in Mixed Infections in Argentina
by
Julia M. Ibañez, Romina Zambrana, Pamela Carreras, Verónica Obregón, José M. Irazoqui, Pablo A. Vera, Tatiana E. Lattar, María D. Blanco Fernández, Andrea F. Puebla, Ariel F. Amadio, Carolina Torres and Paola M. López Lambertini
Viruses 2025, 17(4), 533; https://doi.org/10.3390/v17040533 (registering DOI) - 5 Apr 2025
Abstract
Tobamovirus fructirugosum (ToBRFV) and Crinivirus tomatichlorosis (ToCV) are emerging viral threats to tomato production worldwide, with expanding global distribution. Both viruses exhibit distinct biological characteristics and transmission mechanisms that influence their spread. This study aimed to reconstruct the complete genomes of ToBRFV and
[...] Read more.
Tobamovirus fructirugosum (ToBRFV) and Crinivirus tomatichlorosis (ToCV) are emerging viral threats to tomato production worldwide, with expanding global distribution. Both viruses exhibit distinct biological characteristics and transmission mechanisms that influence their spread. This study aimed to reconstruct the complete genomes of ToBRFV and ToCV from infected tomato plants and wastewater samples in Argentina to explore their global evolutionary dynamics. Additionally, it compared the genetic diversity of ToBRFV in plant tissue and sewage samples. Using metagenomic analysis, the complete genome sequences of two ToBRFV isolates and two ToCV isolates from co-infected tomatoes, along with four ToBRFV isolates from sewage, were obtained. The analysis showed that ToBRFV exhibited higher genetic diversity in environmental samples than in plant samples. Phylodynamic analysis indicated that both viruses had a recent, single introduction in Argentina but predicted different times for ancestral diversification. The evolutionary analysis estimated that ToBRFV began its global diversification in June 2013 in Israel, with rapid diversification and exponential growth until 2020, after which the effective population size declined. Moreover, ToCV’s global expansion was characterized by exponential growth from 1979 to 2010, with Turkey identified as the most probable location with the current data available. This study highlights how sequencing and monitoring plant viruses can enhance our understanding of their global spread and impact on agriculture.
Full article
(This article belongs to the Section Viruses of Plants, Fungi and Protozoa)
►▼
Show Figures

Figure 1
Open AccessCommunication
Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA
by
Anushi Suwaneththiya Deraniyagala, Avijit Roy, Shyam Tallury, Hari Kishan Sudini, Albert K. Culbreath and Sudeep Bag
Viruses 2025, 17(4), 532; https://doi.org/10.3390/v17040532 (registering DOI) - 5 Apr 2025
Abstract
Groundnut bud necrosis orthotospovirus (GBNV), a tripartite single-stranded RNA virus, poses a significant threat to United States agriculture. GBNV is a quarantine pathogen, and its introduction could lead to severe damage to economically important crops, such as groundnuts, tomatoes, potatoes, peas, and soybeans.
[...] Read more.
Groundnut bud necrosis orthotospovirus (GBNV), a tripartite single-stranded RNA virus, poses a significant threat to United States agriculture. GBNV is a quarantine pathogen, and its introduction could lead to severe damage to economically important crops, such as groundnuts, tomatoes, potatoes, peas, and soybeans. For the rapid and accurate detection of GBNV at points of entry, TaqMan reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR) assays were developed and the results validated using reverse transcriptase–polymerase chain reaction (RT-PCR) followed by Sanger sequencing. These assays target highly conserved regions of the nucleocapsid (NP) and movement (MP) proteins within the viral genome. Multiplex GBNV detection assays targeting the NP and MP genes, as well as an internal control gene, ACT11, showed efficiency rates between 90% and 100% and R2 values of 0.98 to 0.99, indicating high accuracy and precision. Moreover, there was no significant difference in sensitivity between multiplex and singleplex assays, ensuring reliable detection across various plant tissues. This rapid, sensitive, and specific diagnostic assay will provide a valuable tool at ports of entry to prevent the entry of GBNV into the United States.
Full article
(This article belongs to the Special Issue Emerging and Reemerging Plant Viruses in a Changing World)
►▼
Show Figures

Figure 1
Open AccessSystematic Review
Prevalence of HHV-6 Detection Among People Living with HIV: A Systematic Review and Meta-Analysis
by
Georgia Kostare, Evangelos Kostares, Michael Kostares, Perry N. Halkitis, Athanasios Tsakris, Theodoros Xanthos and Maria Kantzanou
Viruses 2025, 17(4), 531; https://doi.org/10.3390/v17040531 (registering DOI) - 5 Apr 2025
Abstract
Human herpesvirus 6 (HHV-6) is a ubiquitous virus with significant implications for immunocompromised individuals, particularly people living with HIV (PLWH). This study aimed to estimate the prevalence of HHV-6 detection in blood samples among PLWH using molecular diagnostic techniques. A systematic literature search
[...] Read more.
Human herpesvirus 6 (HHV-6) is a ubiquitous virus with significant implications for immunocompromised individuals, particularly people living with HIV (PLWH). This study aimed to estimate the prevalence of HHV-6 detection in blood samples among PLWH using molecular diagnostic techniques. A systematic literature search was conducted across multiple databases until September 2024, including studies that reported HHV-6 detection in blood samples of PLWH through molecular methods. The meta-analysis calculated pooled prevalence rates using a random-effects model and assessed study quality, with additional analyses for outlier identification and influential study effects. Twelve studies met the inclusion criteria, and the random-effects model estimated the prevalence of HHV-6 detection at 11.7% (95% CI: 4.3–21.8%), with considerable heterogeneity. Influence diagnostics identified one study as influential, and after its exclusion, the recalculated pooled prevalence was 8% (95% CI: 4.4–12.4%), with reduced but still considerable heterogeneity. This meta-analysis highlights the prevalence of HHV-6 detection in PLWH, emphasizing the need for ongoing research to explore the clinical implications and factors influencing viral detection as well as the implications of this coinfection on the treatment and overall health of PLWH.
Full article
(This article belongs to the Special Issue Viral Infections in Special Populations)
►▼
Show Figures

Figure 1
Open AccessArticle
Sulfatide Binds to Influenza B Virus and Enhances Viral Replication
by
Yuuki Kurebayashi, Yoshiki Wakabayashi, Tadanobu Takahashi, Keiko Sakakibara, Shunsaku Takahashi, Akira Minami, Takashi Suzuki and Hideyuki Takeuchi
Viruses 2025, 17(4), 530; https://doi.org/10.3390/v17040530 (registering DOI) - 5 Apr 2025
Abstract
Seasonal influenza epidemics caused by influenza A viruses (IAV) and influenza B viruses (IBV) pose a substantial public health burden. Despite the significant impact of IBV, its restricted host range and the absence of documented pandemics have resulted in limited research attention relative
[...] Read more.
Seasonal influenza epidemics caused by influenza A viruses (IAV) and influenza B viruses (IBV) pose a substantial public health burden. Despite the significant impact of IBV, its restricted host range and the absence of documented pandemics have resulted in limited research attention relative to IAV. Understanding the viral infection mechanisms of both IAV and IBV is crucial for controlling seasonal epidemics. Previously, we demonstrated that 3′-O-sulfated galactosylceramide sulfatide binds to IAV and enhances viral replication, a finding with potential therapeutic implications. However, the role sulfatide plays in other influenza virus infections, including those caused by IBV, remains unknown. Accordingly, in this paper, we investigate the function of sulfatide during IBV infection. We demonstrate that sulfatide binds to IBV hemagglutinin (HA), and that sulfatide overexpression significantly enhances IBV replication, whereas treatment with sulfatase or an anti-sulfatide antibody markedly suppressed IBV replication. Moreover, further tests involving the inhibition of sulfatide biosynthesis resulted in the suppression of viral replication with impaired nuclear export of viral ribonucleoproteins (vRNPs). These findings establish that sulfatide is a critical regulator of IBV replication, which parallels its role in IAV infection, and suggest that targeting sulfatide-virus interactions can lead to broad-spectrum therapeutic strategies against influenza virus.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures
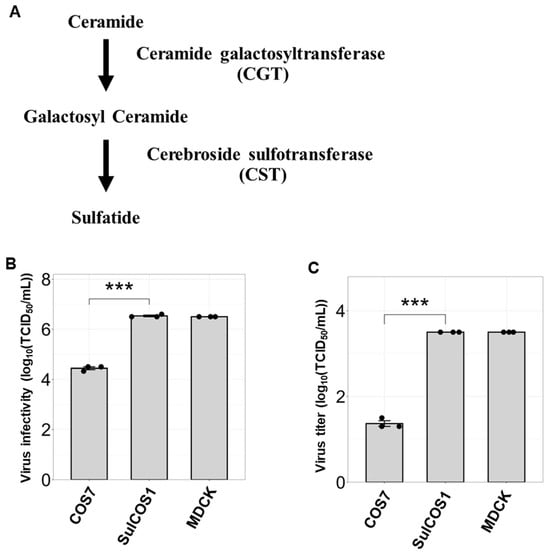
Figure 1
Open AccessArticle
Protective Efficacy of a Chimeric Pestivirus KD26_E2LOM Vaccine Against Classical Swine Fever Virus Infection of Pigs
by
Young-Hyeon Lee, Bo-Kyoung Jung, Song-Yi Kim, Dohyun Kim, Min-Kyung Jang, SeEun Choe, Byung-Hyun An, Jae-Jo Kim, Yun Sang Cho and Dong-Jun An
Viruses 2025, 17(4), 529; https://doi.org/10.3390/v17040529 (registering DOI) - 5 Apr 2025
Abstract
A chimeric pestivirus KD26_E2LOM strain can induce antibodies that can be partially distinguished from antibodies from classical swine fever virus (CSFV) infection. The chimeric pestivirus vaccine strain was created using bovine viral diarrhea virus as the backbone; however, the entire BVDV E2 gene
[...] Read more.
A chimeric pestivirus KD26_E2LOM strain can induce antibodies that can be partially distinguished from antibodies from classical swine fever virus (CSFV) infection. The chimeric pestivirus vaccine strain was created using bovine viral diarrhea virus as the backbone; however, the entire BVDV E2 gene region was replaced with the E2 gene, which encodes the major target for neutralizing antibodies against CSFV. Pigs were vaccinated once or twice with the chimeric pestivirus KD26_E2LOM strain, and protective efficacy was evaluated after subsequent challenge with virulent CSFV. Pigs inoculated with the chimeric pestivirus KD26_E2LOM strain did not have a high temperature or leukopenia, and CSFV neutralizing antibodies (>64-fold) were observed from 28 days postvaccination (dpv). In addition, the level of anti-CSFV E2 antibody positivity was >0.8 (s/p value) from 30 dpv, and there were no antibody-positive individuals among the sentinel pigs. In control pigs, CSF antigen was detected in blood, nasal, and fecal samples at 5, 7, 10, 14, and 21 days postchallenge (dpc) and in several organs; however, no CSFV was detected in the organs of pigs vaccinated with the chimeric pestivirus KD26_E2LOM strain, and no virus shedding or CSF antigen was detected on any dpc. Thus, the chimeric pestivirus KD26_E2LOM strain protects pigs against horizontal transmission of virulent CSFV; however, this strain may have only partial potential for the differential detection of CSFV Erns antibodies.
Full article
(This article belongs to the Special Issue Pestivirus 2025)
►▼
Show Figures

Figure 1
Open AccessReview
Chasing Virus Replication and Infection: PAMP-PRR Interaction Drives Type I Interferon Production, Which in Turn Activates ISG Expression and ISGylation
by
Imaan Muhammad, Kaia Contes, Moses T. Bility and Qiyi Tang
Viruses 2025, 17(4), 528; https://doi.org/10.3390/v17040528 - 4 Apr 2025
Abstract
The innate immune response, particularly the interferon-mediated pathway, serves as the first line of defense against viral infections. During virus infection, viral pathogen-associated molecular patterns (PAMPs) are recognized by host pattern recognition receptors (PRRs), triggering downstream signaling pathways. This leads to the activation
[...] Read more.
The innate immune response, particularly the interferon-mediated pathway, serves as the first line of defense against viral infections. During virus infection, viral pathogen-associated molecular patterns (PAMPs) are recognized by host pattern recognition receptors (PRRs), triggering downstream signaling pathways. This leads to the activation of transcription factors like IRF3, IRF7, and NF-κB, which translocate to the nucleus and induce the production of type I interferons (IFN-α and IFN-β). Once secreted, type I interferons bind to their receptors (IFNARs) on the surfaces of infected and neighboring cells, activating the JAK-STAT pathway. This results in the formation of the ISGF3 complex (composed of STAT1, STAT2, and IRF9), which translocates to the nucleus and drives the expression of interferon-stimulated genes (ISGs). Some ISGs exert antiviral effects by directly or indirectly blocking infection and replication. Among these ISGs, ISG15 plays a crucial role in the ISGylation process, a ubiquitin-like modification that tags viral and host proteins, regulating immune responses and inhibiting viral replication. However, viruses have evolved counteractive strategies to evade ISG15-mediated immunity and ISGylation. This review first outlines the PAMP-PRR-induced pathways leading to the production of cytokines and ISGs, followed by a summary of ISGylation’s role in antiviral defense and viral evasion mechanisms targeting ISG15 and ISGYlation.
Full article
(This article belongs to the Special Issue Molecular Epidemiology, Evolution, and Dispersion of Flaviviruses (2nd Edition))
►▼
Show Figures

Figure 1
Open AccessArticle
Genetic and Phenotypic Investigations of Viral Subpopulations Detected in Different Tissues of Laying Hens Following Infectious Bronchitis Virus Infection
by
Ahmed Ali, Ryan Rahimi, Motamed Elsayed Mahmoud, Adel A. Shalaby, Rodrigo A. Gallardo and Mohamed Faizal Abdul-Careem
Viruses 2025, 17(4), 527; https://doi.org/10.3390/v17040527 - 4 Apr 2025
Abstract
Infectious bronchitis virus (IBV) commonly produces a range of genetic sequences during replication, particularly in the spike 1 (S1)-coding portion of the S gene, leading to distinct subpopulations within the broader viral population. It has been shown that certain microenvironments exert selective pressure
[...] Read more.
Infectious bronchitis virus (IBV) commonly produces a range of genetic sequences during replication, particularly in the spike 1 (S1)-coding portion of the S gene, leading to distinct subpopulations within the broader viral population. It has been shown that certain microenvironments exert selective pressure on the S1-coding sequences and their encoded proteins, influencing the selection of viral subpopulations in these environments. In this study, high-throughput next-generation sequencing (NGS) was used to analyze the S1-coding sequences from tissues of the respiratory, digestive, renal, and reproductive systems of specific pathogen-free (SPF) laying hens. These tissues were collected nine days after infection with the California 1737/04 (CA1737/04) IBV strain, which is known to cause varying degrees of pathology in these tissues. Using a specific bioinformatics pipeline, 27 single nucleotide variants (SNVs) were detected in the S1-coding sequences derived from different tissues. These SNVs shaped multiple subpopulations (SP1–SP15), with SP1 being the core subpopulation present in all tissues, while others were tissue-specific. The IBV RNA loads in the tissues were negatively correlated with the number of SNVs or the Shannon entropy values, and phylogenetic analysis revealed a genetic divergence in the S1-coding sequences from certain tissues with lower viral RNA loads, particularly those from the trachea and ovary. Furthermore, the SNVs were associated with nonsynonymous mutations, primarily located in hypervariable region 2 (HVR 2) within the N-terminal domain of S1 (S1-NTD), except for those in SP7, which was exclusive to the trachea and contained changes in HVR 3 in the C-terminal domain of S1 (S1-CTD). Overall, this study adds to the existing knowledge about IBV evolution by highlighting the role of tissue-specific environments in shaping viral genetic diversity.
Full article
(This article belongs to the Special Issue Enteric and Respiratory Viruses in Animals and Birds: Volume 5)
►▼
Show Figures

Figure 1
Open AccessArticle
The Ongoing Epidemics of Seasonal Influenza A(H3N2) in Hangzhou, China, and Its Viral Genetic Diversity
by
Xueling Zheng, Feifei Cao, Yue Yu, Xinfen Yu, Yinyan Zhou, Shi Cheng, Xiaofeng Qiu, Lijiao Ao, Xuhui Yang, Zhou Sun and Jun Li
Viruses 2025, 17(4), 526; https://doi.org/10.3390/v17040526 - 4 Apr 2025
Abstract
This study examined the genetic and evolutionary features of influenza A/H3N2 viruses in Hangzhou (2010–2022) by analyzing 28,651 influenza-like illness samples from two sentinel hospitals. Influenza A/H3N2 coexisted with other subtypes, dominating seasonal peaks (notably summer). Whole-genome sequencing of 367 strains was performed
[...] Read more.
This study examined the genetic and evolutionary features of influenza A/H3N2 viruses in Hangzhou (2010–2022) by analyzing 28,651 influenza-like illness samples from two sentinel hospitals. Influenza A/H3N2 coexisted with other subtypes, dominating seasonal peaks (notably summer). Whole-genome sequencing of 367 strains was performed on GridION platforms. Phylogenetic analysis showed they fell into 16 genetic groups, with multiple clades circulating simultaneously. Shannon entropy indicated HA, NA, and NS gene segments exhibited significantly higher variability than other genomic segments, with HA glycoprotein mutations concentrated in antigenic epitopes A–E. Antiviral resistance showed no inhibitor resistance mutations in PA, PB1, or PB2, but NA mutations were detected in some strains, and most strains harbored M2 mutations. A Bayesian molecular clock showed the HA segment exhibited the highest nucleotide substitution rate (3.96 × 10−3 substitutions/site/year), followed by NA (3.77 × 10−3) and NS (3.65 × 10−3). Selective pressure showed A/H3N2 strains were predominantly under purifying selection, with only sporadic positive selection at specific sites. The Pepitope model demonstrated that antigenic epitope mismatches between circulating H3N2 variants and vaccine strains led to a significant decline in influenza vaccine effectiveness (VE), particularly in 2022. Overall, the study underscores the complex circulation patterns of influenza in Hangzhou and the global importance of timely vaccine strain updates.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
Modeling and Molecular Dynamics Studies of Flavone―DENV E-3 Protein―SWCNT Interaction at the Flavonoid Binding Sites
by
Cecilia Espíndola
Viruses 2025, 17(4), 525; https://doi.org/10.3390/v17040525 - 4 Apr 2025
Abstract
The DENV virus circulates freely in endemic regions and causes dengue disease. The vectors are Aedes aegypti and Aedes albopictus. The difficulties inherent in the nature of the DENV virus, its epidemiology, and its increasing incidence in recent years have led to
[...] Read more.
The DENV virus circulates freely in endemic regions and causes dengue disease. The vectors are Aedes aegypti and Aedes albopictus. The difficulties inherent in the nature of the DENV virus, its epidemiology, and its increasing incidence in recent years have led to the development of viable alternatives in the search for effective solutions for the treatment of this severe disease. Flavones such as tropoflavin, baicalein, and luteolin have anti-DENV activity. Molecular docking studies were performed between the flavones tropoflavin, baicalein, and luteolin and the DENV E-3 protein. Flavone—DENV E-3 complex interactions were analyzed at the flavonoid binding sites domain I of the B chain and domain II of the A chain reported in the literature. H-bond, π-π stacking, and π-cation interactions between flavones and the DENV E-3 protein at different binding energies were evaluated. Molecular dynamics studies for these interactions were performed to determine the molecular stability of the Flavone—DENV E-3 complexes. I also present here the results of the molecular interactions of the Flavone—DENV E-3―SWCNT complex. Due to recent advances in nanotechnology and their physicochemical properties, the utilization of nanoparticles such as SWCNT has increased in antiviral drug delivery.
Full article
(This article belongs to the Special Issue Novel Antiviral Agents: Synthesis, Molecular Modelling Studies and Biological Investigation, 2nd Edition)
►▼
Show Figures

Figure 1
Open AccessCommunication
Recent Hepatitis E Virus Infection in Wild Boars and Other Ungulates in Japan
by
Milagros Virhuez-Mendoza, Keita Ishijima, Kango Tatemoto, Yudai Kuroda, Yusuke Inoue, Ayano Nishino, Tsukasa Yamamoto, Akihiko Uda, Akitoyo Hotta, Hidenori Kabeya, Hiroshi Shimoda, Kazuo Suzuki, Tomoyoshi Komiya, Junji Seto, Yuki Iwashina, Daisuke Hirano, Mikio Sawada, Sayuri Yamaguchi, Fusayo Hosaka and Ken Maeda
Viruses 2025, 17(4), 524; https://doi.org/10.3390/v17040524 - 4 Apr 2025
Abstract
Hepatitis E virus (HEV) is a zoonotic pathogen with multiple hosts, posing significant public health risks, especially in regions like Japan where game meat consumption is prevalent. This study investigated HEV infection and viral shedding in wild boars, sika deer, and Japanese serows
[...] Read more.
Hepatitis E virus (HEV) is a zoonotic pathogen with multiple hosts, posing significant public health risks, especially in regions like Japan where game meat consumption is prevalent. This study investigated HEV infection and viral shedding in wild boars, sika deer, and Japanese serows across Japan. A total of 1896 serum samples were tested for anti-HEV antibodies, 1034 for HEV RNA, and 473 fecal samples for viral shedding. Anti-HEV antibodies were detected in wild boars from all seven prefectures studied, while HEV RNA was detected in wild boars from Fukuoka, Oita, and Miyazaki in southern Japan, as well as Yamaguchi prefecture. Genetic analysis revealed subtypes 3b, 4a, and 4g, with 3b being the most prevalent. Subtype 3b exhibited distinct geographical clustering, whereas 4g persisted exclusively in Yamaguchi for over 12 years. Infectious HEV particles were confirmed in wild boar feces, highlighting the risk of environmental contamination and zoonotic transmission. Sika deer showed no evidence of HEV infection, and only one Japanese serow tested positive for antibodies without detectable RNA. These findings underscore the importance of ongoing surveillance to assess the zoonotic risks from game meat consumption and prevention of HEV transmission to humans.
Full article
(This article belongs to the Special Issue Hepatitis E: Molecular Virology, Pathogenesis, and Treatment, 2nd Edition)
►▼
Show Figures

Figure 1
Open AccessArticle
Compelling Increase in Parvovirus B19 Infections: Analysis of Molecular Diagnostic Trends (2019–2024)
by
Flora Marzia Liotti, Simona Marchetti, Sara D’Onghia, Lucio Romano, Rosalba Ricci, Maurizio Sanguinetti, Rosaria Santangelo and Brunella Posteraro
Viruses 2025, 17(4), 523; https://doi.org/10.3390/v17040523 - 4 Apr 2025
Abstract
Human parvovirus B19 (B19V) follows a well-documented cyclical epidemiology, with peaks occurring every 3–4 years. However, recent reports indicate an unusual resurgence in B19V infections across multiple countries, prompting increased surveillance. This study analyzed molecular diagnostic assay results from 826 unique-patient samples tested
[...] Read more.
Human parvovirus B19 (B19V) follows a well-documented cyclical epidemiology, with peaks occurring every 3–4 years. However, recent reports indicate an unusual resurgence in B19V infections across multiple countries, prompting increased surveillance. This study analyzed molecular diagnostic assay results from 826 unique-patient samples tested for B19V DNA between 2019 and 2024 at a large Italian tertiary-care hospital, covering pre-, during, and post-COVID-19 years. Overall, 80 of 826 patients (9.7%) tested positive for B19V DNA. A significant increase in positivity was observed in 2024 (23.4%), with a peak in May, representing an eightfold rise compared to 2019–2020. Despite this surge, the distribution of positive cases across population categories remained consistent with previous years, with 32 of 80 (40.0%) positive samples from pregnant women and 27 of 80 (33.8%) from hematology/oncology patients. Among 66 B19V DNA-positive patients with available serology, 4 of 66 (6.1%)—all immunocompromised—lacked detectable IgM/IgG despite high B19V DNA levels (7.8 log10 IU/mL). These findings highlight the importance of integrating molecular and serological diagnostics, particularly in high-risk populations. Given the potential impact of the COVID-19 pandemic on B19V circulation, continued surveillance is essential to determine whether this resurgence represents a temporary fluctuation or a sustained epidemiological shift.
Full article
(This article belongs to the Collection Parvoviridae)
►▼
Show Figures

Figure 1
Open AccessArticle
The Role of the Tyrosine-Based Sorting Signals of the ORF3a Protein of SARS-CoV-2 in Intracellular Trafficking and Pathogenesis
by
Edward B. Stephens, Dusan Kunec, Wyatt Henke, Ricardo Martin Vidal, Brandon Greishaber, Rabina Saud, Maria Kalamvoki, Gagandeep Singh, Sujan Kafle, Jessie D. Trujillo, Franco Matias Ferreyra, Igor Morozov and Juergen A. Richt
Viruses 2025, 17(4), 522; https://doi.org/10.3390/v17040522 - 3 Apr 2025
Abstract
The open reading frame 3a (ORF3a) is a protein important to the pathogenicity of SARS-CoV-2. The cytoplasmic domain of ORF3a has three canonical tyrosine-based sorting signals (160YNSV163, 211YYQL213, and 233YNKI236), and a previous study has indicated that mutation of the 160YNSV163 motif abrogated
[...] Read more.
The open reading frame 3a (ORF3a) is a protein important to the pathogenicity of SARS-CoV-2. The cytoplasmic domain of ORF3a has three canonical tyrosine-based sorting signals (160YNSV163, 211YYQL213, and 233YNKI236), and a previous study has indicated that mutation of the 160YNSV163 motif abrogated plasma membrane expression and inhibited ORF3a-induced apoptosis. Here, we have systematically removed all three tyrosine-based motifs and assessed the importance of each motif or combination of motifs in trafficking to the cell surface. Our results indicate that the 160YNSV163 motif alone was insufficient for ORF3a cell-surface trafficking, while the 211YYQL213 motif was the most important. Additionally, an ORF3a with all three YxxΦ motifs disrupted (ORF3a-[ΔYxxΦ]) was not transported to the cell surface, and LysoIP studies indicate that ORF3a but not ORF3a-[ΔYxxΦ] was present in late endosome/lysosome fractions. A growth-curve analysis of different SARS-CoV-2 viruses expressing the different mutant ORF3a proteins revealed no significant differences in virus replication. Finally, the inoculation of K18hACE-2 mice indicated that the SARS-CoV-2 lacking the three YxxΦ motifs was less pathogenic than the unmodified SARS-CoV-2. These results indicate that the tyrosine motifs of ORF3a contribute to cell-surface expression and SARS-CoV-2 pathogenesis
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
Impact of BTV-3 Circulation in Belgium in 2024 and Current Knowledge Gaps Hindering an Evidence-Based Control Program
by
Virginie Van Leeuw, Ilse De Leeuw, Nicolas Degives, Pieter Depoorter, Jeroen Dewulf, Jean-Baptiste Hanon, Jozef Hooyberghs, Annick Linden, Laura Praet, Marc Raemaekers, Claude Saegerman, Xavier Simons, Charlotte Sohier, Norbert Steurbaut, Amandine Sury, Etienne Thiry, Stephan Zientara, Axel Mauroy and Nick De Regge
Viruses 2025, 17(4), 521; https://doi.org/10.3390/v17040521 - 3 Apr 2025
Abstract
Between 2006 and 2010, northwestern Europe experienced its first significant bluetongue virus (BTV) outbreak, driven by the spread of BTV-8, which had major repercussions on the European livestock sector. While BTV-3 was first identified in Europe in Italy in 2017, a new introduction
[...] Read more.
Between 2006 and 2010, northwestern Europe experienced its first significant bluetongue virus (BTV) outbreak, driven by the spread of BTV-8, which had major repercussions on the European livestock sector. While BTV-3 was first identified in Europe in Italy in 2017, a new introduction of the virus was reported in 2023, in the Netherlands, and subsequently spread rapidly across the continent. A limited number of BTV-3 outbreaks were notified in Belgium in 2023, leading to the loss of its BTV-free status. In the following year, 2024, the virus spread throughout the country in a short time period. This study describes the impact of BTV-3 circulation in Belgium in 2024, detailing both its geographic spread and the associated increase in mortality, reduced births recorded, and decline in milk production among ruminants. Furthermore, preliminary results on the effectiveness of field vaccination and maternal immunity transfer are presented, as well as critical gaps that hinder the development of a robust, evidence-based management strategy. As the epidemiological situation is expected to become more complex in the future, due to the co-circulation of multiple BTV serotypes and other Culicoides-borne diseases, such as EHDV, effective collaboration and communication among stakeholders and international authorities will be crucial for implementing measures to mitigate the spread of these diseases.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
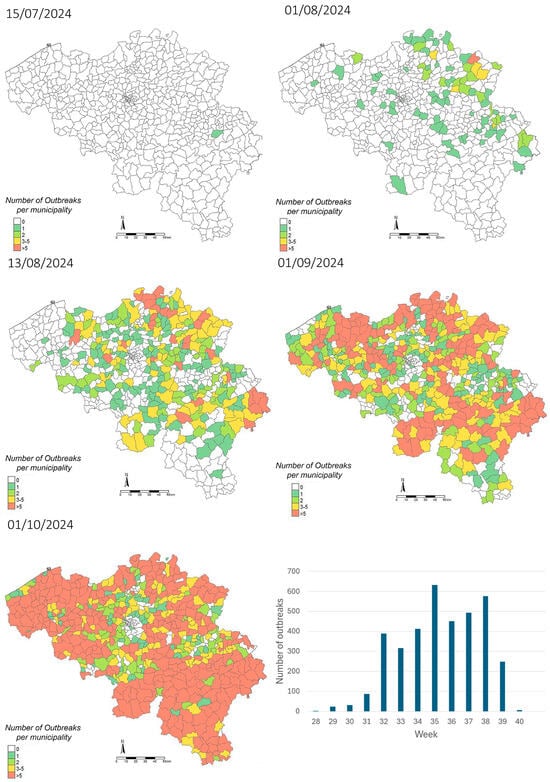
Figure 1
Open AccessArticle
Relationship Between Gut Microbiota and the Clinical Course of COVID-19 Disease
by
Antonija Jonjić, Ivan Dolanc, Goran Slivšek, Luka Bočkor, Marko Tarle, Sanda Mustapić, Marta Kmet, Biserka Orehovec, Paola Kučan Brlić, Maja Cokarić Brdovčak, Ante Obad, Martin Walenta, Ivan Dražić, Lidija Bilić-Zulle, Ivica Lukšić, Neven Bulić, Walter Goessler, Stipan Jonjić, Miran Čoklo and Jurica Žučko
Viruses 2025, 17(4), 520; https://doi.org/10.3390/v17040520 - 2 Apr 2025
Abstract
Possible early detection of people at increased risk for severe COVID-19 clinical course is extremely important so that appropriate therapy can be initiated promptly to prevent numerous deaths. Our study included 45 patients treated for COVID-19 at Dubrava University Hospital, with clinical course
[...] Read more.
Possible early detection of people at increased risk for severe COVID-19 clinical course is extremely important so that appropriate therapy can be initiated promptly to prevent numerous deaths. Our study included 45 patients treated for COVID-19 at Dubrava University Hospital, with clinical course analysed from medical records and stool samples collected for determination of the gut microbiota diversity using 16S rRNA analysis. Sequencing was successful for 41 samples belonging to four clinical course groups (WHO guidelines): 12 samples—critical, 12—severe, 9—moderate and 8—mild group. Microbial composition was assessed between groups using two approaches—ANCOM (QIIME2) and Kruskal–Wallis (MicrobiomeAnalyst). On the genus level, two taxa were found to be differentially abundant: archaeal Halococcus and Coprococcus (for both W = 37)—the two were most abundant in the critical group (10% and 0.94% of entire abundance, respectively). Coprococcus catus was the only species identified by both methods to be differentially abundant between groups and was most abundant in the critical group. Alpha diversity indicated greater evenness of features in the critical group. Beta diversity showed clustering of samples from the critical group. A relationship between gut microbiota composition and the clinical course of COVID-19 disease was indicated, pointing towards specific distinct features of the critical group. In a broader sense, our findings might be useful in combating potential future similar pandemics and emerging virus outbreaks.
Full article
(This article belongs to the Special Issue Advanced Strategies against SARS-CoV-2 Variants and Future Emerging Virus Outbreaks)
►▼
Show Figures

Figure 1
Open AccessCommunication
Diagnostic Findings of Transmissible Viral Proventriculitis Associated with Chicken Proventricular Necrosis Virus in Processed Broiler Chickens in Argentina
by
Carlos Daniel Gornatti-Churria, Natàlia Majó, Melissa Macías-Rioseco, Rosa M. Valle, Patricio A. García and Carmen F. Jerry
Viruses 2025, 17(4), 519; https://doi.org/10.3390/v17040519 - 1 Apr 2025
Abstract
Transmissible viral proventriculitis (TVP) and chicken proventricular necrosis virus (CPNV) affect the broiler industry globally and are emerging diseases of economic importance. Here, we present the findings of TVP from processed broiler carcasses in Argentina following marked condemnation at the processing plant. We
[...] Read more.
Transmissible viral proventriculitis (TVP) and chicken proventricular necrosis virus (CPNV) affect the broiler industry globally and are emerging diseases of economic importance. Here, we present the findings of TVP from processed broiler carcasses in Argentina following marked condemnation at the processing plant. We studied a total of 122 abnormally presenting proventriculi at processing from 42-to-50-day-old, male Cobb500™ broiler chicken carcasses from 11 farms belonging to the same company in 13 episodes of proventriculi–gizzards condemnation between December 2021 and April 2022. The proventriculi were enlarged and pale with a widened gastric isthmus. A histopathologic lesion score system was developed based on the presence of a combination of key microscopic findings, the distribution, and the severity of the lesions. Scoring of the affected proventriculi revealed 65% (79/122) with a score of 4, 23% (28/122) with a score of 3, and 12% (15/122) with a score of 2. Focal to multifocal immunoreactivity against the VP2-CPNV antigen within the necrotic glandular epithelial cells was noted in the affected proventriculi using immunohistochemistry. We found 84.4% (103/122) of the studied proventriculi with TVP lesions grossly and microscopically scored were positive for CPNV by RT-PCR. The sequencing results of the PCR product showed a high nucleotide sequence similarity (88.97%) to previously published VP1-CPNV sequences. We confirmed CPNV infection in most of the TVP affected proventriculi in all condemnation episodes at a broiler chicken processing plant in Argentina during the studied period. This study documents TVP associated with CPNV detection at processing plants in Argentina for the first time.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
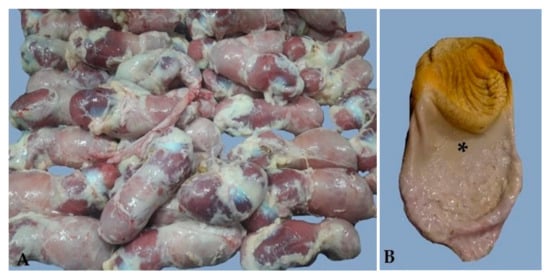
Figure 1
Open AccessArticle
Nanotechnology-Driven Strategy Against SARS-CoV-2: Pluronic F127-Based Nanomicelles with or Without Atazanavir Reduce Viral Replication in Calu-3 Cells
by
Eduardo Ricci-Junior, Alice Santos Rosa, Tatielle do Nascimento, Ralph Santos-Oliveira, Marcos Alexandre Nunes da Silva, Debora Ferreira Barreto-Vieira, Luísa Tozatto Batista, Giovanna Barbosa da Conceição, Tayane Alvites Nunes Quintão, Vivian Neuza Santos Ferreira and Milene Dias Miranda
Viruses 2025, 17(4), 518; https://doi.org/10.3390/v17040518 - 1 Apr 2025
Abstract
Despite extensive efforts, no highly effective antiviral molecule exists for treating moderate and severe COVID-19. Nanotechnology has emerged as a promising approach for developing novel drug delivery systems to enhance antiviral efficacy. Among these, polymeric nanomicelles improve the solubility, bioavailability, and cellular uptake
[...] Read more.
Despite extensive efforts, no highly effective antiviral molecule exists for treating moderate and severe COVID-19. Nanotechnology has emerged as a promising approach for developing novel drug delivery systems to enhance antiviral efficacy. Among these, polymeric nanomicelles improve the solubility, bioavailability, and cellular uptake of therapeutic agents. In this study, Pluronic F127-based nanomicelles were developed and evaluated for their antiviral activity against SARS-CoV-2. The nanomicelles, formulated using the direct dissolution method, exhibited an average size of 37.4 ± 8.01 nm and a polydispersity index (PDI) of 0.427 ± 0.01. Their antiviral efficacy was assessed in SARS-CoV-2-infected Vero E6 and Calu-3 cell models, where treatment with a 1:2 dilution inhibited viral replication by more than 90%. Cytotoxicity assays confirmed the nanomicelles were non-toxic to both cell lines after 72 h. In SARS-CoV-2-infected Calu-3 cells (human type II pneumocyte model), treatment with Pluronic F127-based nanomicelles containing atazanavir (ATV) significantly reduced viral replication, even under high MOI (2) and after 48 h, while also preventing IL-6 upregulation. To investigate their mechanism, viral pretreatment with nanomicelles showed no inhibitory effect. However, pre-exposure of Calu-3 cells led to significant viral replication reduction (>85% and >75% for 1:2 and 1:4 dilutions, respectively), as confirmed by transmission electron microscopy. These findings highlight Pluronic F127-based nanomicelles as a promising nanotechnology-driven strategy against SARS-CoV-2, reinforcing their potential for future antiviral therapies.
Full article
(This article belongs to the Special Issue Nanovaccines against Viral Infection)
►▼
Show Figures

Figure 1

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
30 October–2 November 2025
The 11th Wuhan International Symposium on Modern Virology & Viruses 2025 Conference

Special Issues
Special Issue in
Viruses
Epigenetic Modifications in Viral Infections, Volume II
Guest Editors: Matloob Husain, Farjana AhmedDeadline: 15 April 2025
Special Issue in
Viruses
Molecular Biomarkers for Viral Infection
Guest Editor: Paulina Niedźwiedzka-RystwejDeadline: 15 April 2025
Special Issue in
Viruses
Innate Immunity to Virus Infection 2nd Edition
Guest Editors: Caijun Sun, Feng MaDeadline: 15 April 2025
Special Issue in
Viruses
The Challenge of HIV Diversity
Guest Editor: Brian T. FoleyDeadline: 15 April 2025
Topical Collections
Topical Collection in
Viruses
Mathematical Modeling of Viral Infection
Collection Editors: Amber M. Smith, Ruian Ke
Topical Collection in
Viruses
Efficacy and Safety of Antiviral Therapy
Collection Editors: Giordano Madeddu, Andrea De Vito, Agnese Colpani








